Figure 6.
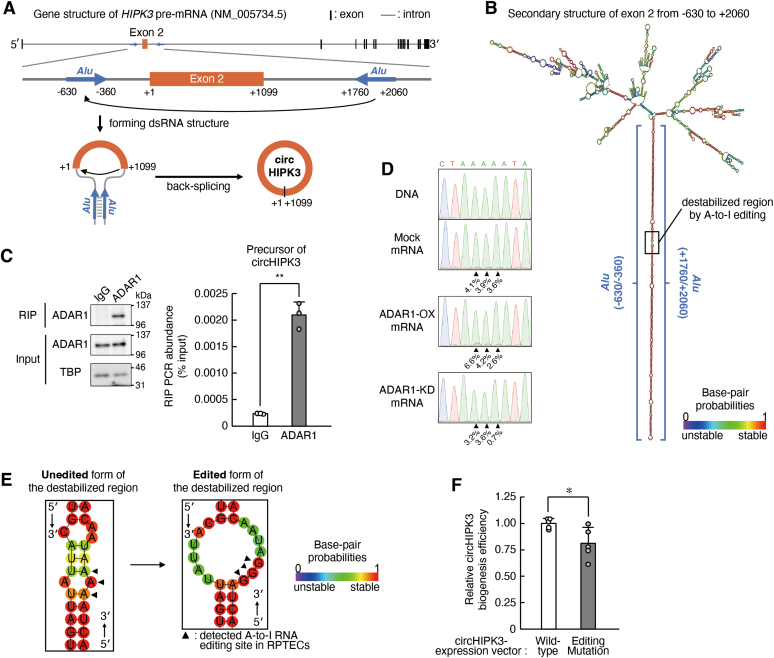
ADAR1 suppresses circHIPK3 formation by inhibiting base pairing in its precursor RNA.A, schematic diagram of the genomic region of human HIPK3 exon 2 and its flanking introns with inverted Alu elements (upper panel). CircHIPK3 is formed by back-splicing, in which a 5′ splice site in the downstream region of exon 2 joins to the 3′ splice site in the upstream region of exon 2 (lower panel). B, in silico analysis for prediction of RNA secondary structure of the HIPK3 pre-mRNA (the region from −630 to +2060 of HIPK3 exon 2) by RNAfold. The minimum free energy structure with base-pair probabilities was calculated to have the lowest value of free energy. Base-pair probabilities are shown by a color spectrum. C, ADAR1 binds to the precursor of circHIPK3. RPTECs were infected with lentiviral particles expressing precursor of circHIPK3 and RIP was conducted. Left panel shows Western blotting analysis of ADAR1-RIP immunoprecipitates. Right panel shows qRT-PCR analysis of ADAR1-RIP immunoprecipitates using pre-circHIPK3 targeting primers listed in Table 1. Values are the mean with SD (n = 3). ∗∗p < 0.01; significant difference between the two groups (t4 = 13.102, p < 0.001; unpaired t test, two sided). D, electropherograms from direct sequencing of the PCR-amplified product of precursor of circHIPK3 derived from lentivirus. The sequence data were obtained from mock-transduced, ADAR1-overexpression (OX), and ADAR1-KD RPTECs. Triangles indicate the ADAR1-mediated A-to-I RNA editing sites detected in RPTECs. Percentage represents the editing frequency calculated by the peak height of “G” peak over sum of “A” and “G” peak heights. Electropherograms were aligned using SnapGene Viewer. E, in silico prediction of RNA secondary structure around the editing sites in precursor of circHIPK3. Left and right panels show the unedited and edited forms, respectively. The minimum free energy structure with base-pair probabilities was calculated to have the lowest value of free energy. Base-pair probabilities are shown by a color spectrum. F, the biogenesis efficiency of circHIPK3 derived from circHIPK3-expression vector with and without the mutation in Alu elements. The efficiency was calculated as the ratio of the expression levels of circHIPK3 to that of precursor of circHIPK3, which were assessed by qRT-PCR analysis. Values are the mean with SD (n = 5). ∗p < 0.05; significant difference between the two groups (t8 = 2.682, p = 0.028; unpaired t test, two sided). RIP, RNA immunoprecipitation; TBP, TATA-binding protein.