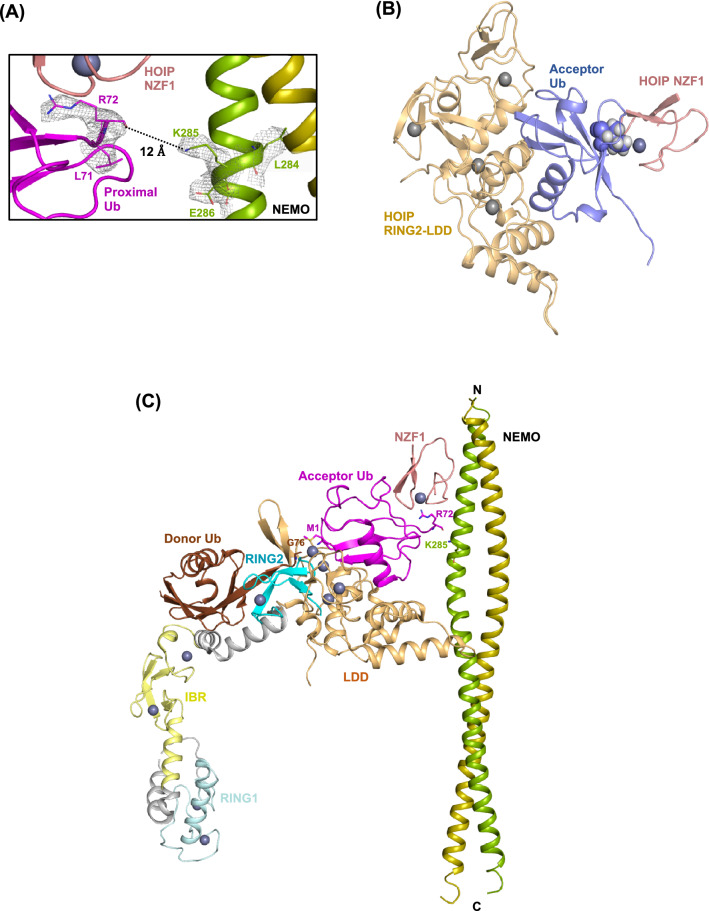
Figure 3.
A structural model for the linear ubiquitination of NEMO by the HOIP E3 ligase. The last four C-terminal amino acid residues of the proximal ubiquitin are not visible in the electron density map of the heteropentameric structure. The distance between the main chain carboxyl group of ubiquitin Arg72 and the sidechain ε-amino group of NEMO Lys285 is ~ 12 Å, which is sufficient to accommodate the last four C-terminal residues of ubiquitin (a.a. 73-76) and form an isopeptide bond. The electron density indicates the 2Fo–Fc map within 1.6 Å of the selected atoms (residues L71 and R72 of ubiquitin and L284-E286 of NEMO), contoured at 1.0 σ (B) Superimposition of the proximal ubiquitin on the acceptor ubiquitin bound to HOIP LDD (PDB ID: 4LJO), suggesting that ubiquitin can interact with NZF1 and RING2-LDD of HOIP. Ile44 residue of ubiquitin molecules are indicated as spheres (C) Superimposition of the three complex structures, including RING2-LDD/acceptor ubiquitin (PDB ID: 4LJO), RBR/donor ubiquitin (PDB ID: 5EDV), and NEMO/NZF1/ubiquitin (current structure) demonstrates a model for the site-specific ligation of linear ubiquitin chains to the NEMO substrate by HOIP. The gray spheres in the figures represent zinc ions.