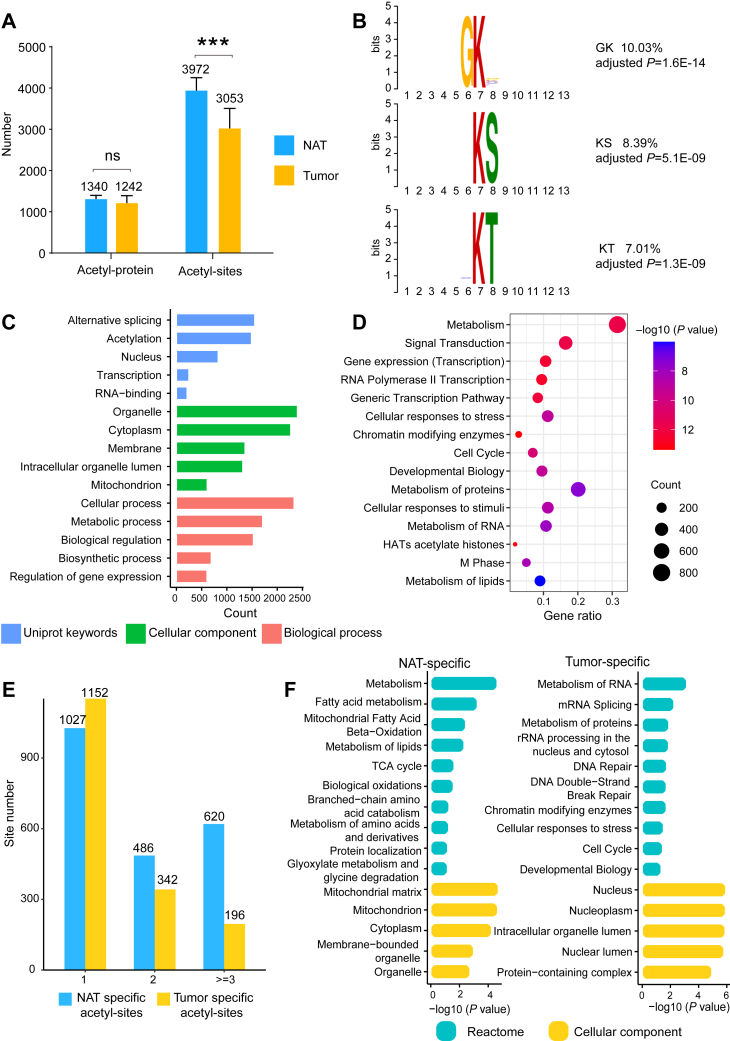
Fig. 3.
Identification of the nonhistone K-acetylation sites in HCC tumors and NATs.A, the average number of K-acetylation sites and the K-acetylated proteins identified in tumor and NAT samples. Error bars represent SDs of eight samples. ns, not significant; ∗∗∗p < 0.001 by two-sided Student’s t-test. B, analysis of the amino acid sequence motifs associated with the detected acetylpeptides. The analysis was conducted by MoMo (version: 5.4.1), using shuffled input peptides as the control. C, GO analysis of the K-acetylated proteins. D, pathway enrichment of the K-acetylated proteins using the Reactome database. E, numbers of the tumor- and NAT-specific K-acetylation sites (n = 1, n = 2, n ≥ 3 out of 8 samples). F, functional enrichment analysis of proteins containing tumor- or NAT-specific acetylation sites (n ≥ 3). The GO and pathway enrichment analysis was conducted by AGOTOOL (https://agotool.org/), using the abundance-corrected proteome as the control for statistical analysis (p < 0.05 as cutoff). See also supplemental Table S4. GO, gene ontology; HCC, hepatocellular carcinoma; NATs, normal adjacent tissues.