Table 1.
The detailed information of the datasets
| Name | Description |
|---|---|
| RefSeq dataset | We split the training and test set by time. All the phage genomes released before December 2018 in RefSeq comprise the training set, while the genomes released after that comprise the test set. This dataset is a widely used benchmark dataset in phage identification task. For each phage, the host information is available based on the keywords 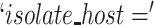 or or 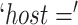 within each GenBank file. If no known host is available, we use this phage as a positive sample without a negative pair. Finally, 305 bacteria and 4410 phages were downloaded. The training set contains 106 bacteria and 2126 phages. The test set contains 194 bacteria and 2284 phages. within each GenBank file. If no known host is available, we use this phage as a positive sample without a negative pair. Finally, 305 bacteria and 4410 phages were downloaded. The training set contains 106 bacteria and 2126 phages. The test set contains 194 bacteria and 2284 phages. |
| Short contig test set | We randomly cut the test phage genomes into segments of different lengths: 1, 2, 3, 5, 10 and 15 kbp. To balance the dataset, we randomly extract 10 segments from each test phage genome and 100 segments from each test bacterial genome. Finally, we have 22 840 phage segments and 19 400 bacterial segments for each given length. Then we use these segments to evaluate the performance of phage identification on short contigs. |
| Simulated metagenomic dataset | We use a sophisticated metagenomic simulator, CAMISIM [40], to generate simulated data using six common bacteria living in the human gut. Instead of adding random phages to this dataset, we add simulated reads from phages that infect these bacteria to create a harder case for distinguishing phages from bacteria with shared local similarities. Then, metaSPAdes [41] is applied to assemble the reads into contigs, which are fed into test phage detection tools. Finally, MetaQUAST [42] is used to map contigs to reference phage genomes in order to assign the labels to the contigs. The experimental results can be found in section Experiments on the simulated data in the [Supplementary file 1]. |
| Mock metagenomic dataset | Nine shotgun metagenomic sequencing replicates of a mock community [43] are retrieved from the European Nucleotide Archive (BioProject PRJEB19901). We use metaSPAdes to assemble the reads into contigs, which are used for evaluation. Similarly, The label of the contigs is determined using MetaQUAST. |
| IMG/VR dataset | IMG/VR v3 database [12] contains 2314 129 viral contigs assembled from different environmental samples. We recruit 354 501 contigs with known bacterial hosts. With this dataset, we will compare the recall of different tools for identifying phages from different environments. |
