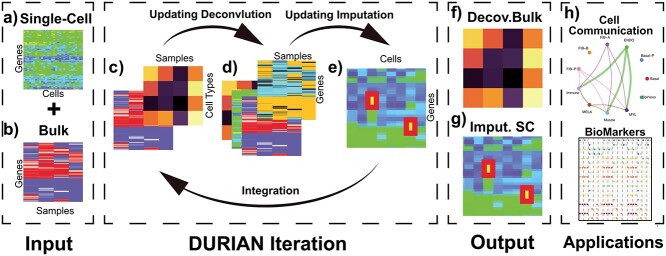
Figure 1.
Schematic of DURIAN method. (A) Input: single-cell expression data (m × n). (B) Input: bulk expression data (m × w). (C) Deconvolution of bulk data, yielding a matrix of celltype proportions (w × k). (D) Construct deconvolution map (m × w) where each column corresponds to the pseudobulk average of the current imputation result according to the celltype ratios calculated in (C). (E) Update the imputation objective with the current deconvolution map. Perform imputation to replace zeros in the single-cell expression data (m × n) with estimated counts. Finally, update the deconvolution model with the imputation estimate. (F) Output: final deconvolution matrix of celltype proportions (w × k). (G) Output: final estimate of single-cell data with dropout replaced by imputed reads. (H) Application: use imputed single-cell data to directly compute cell-signaling patterns.