Figure 3.
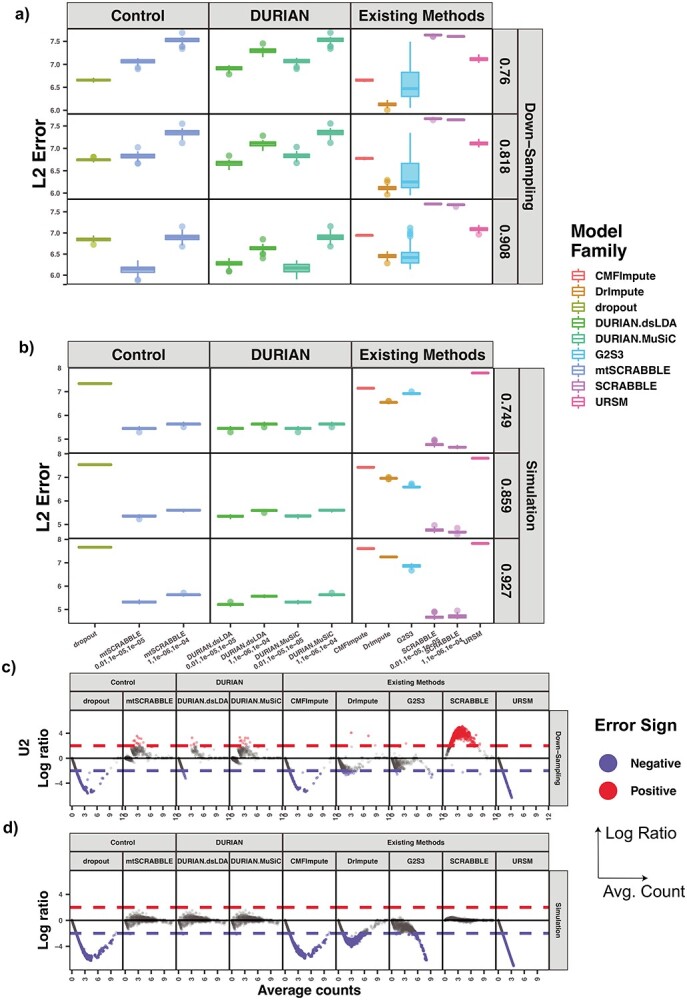
L2 error of Imputation Benchmarks. (A-B) The y-axis is the log L2 norm of difference between the imputed data and the true data, for both synthetic data strategies: downsampling and simulation. For each strategy, the mean sparsity (percentage of zeros in the unimputed data) of all replicates corresponding to each of three values for the strategy-specific dropout parameter is shown in the 2nd level vertical label: 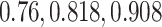 (downsampling),
(downsampling), 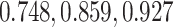 (simulation). The x-axis is arranged into three method categories (shown in top label): Control, DURIAN, Existing Methods. Control methods include dropout: unimputed data, and mtSCRABBLE: the DURIAN algorithm where the deconvolution map is permanently set according to the true bulk celltype percentages. Both dsLDA and NNLS (MuSiC) deconvolution approaches are included for DURIAN benchmarks. Two sets of values for the SCRABBLE objective are provided for DURIAN, SCRABBLE and mtSCRABBLE:
(simulation). The x-axis is arranged into three method categories (shown in top label): Control, DURIAN, Existing Methods. Control methods include dropout: unimputed data, and mtSCRABBLE: the DURIAN algorithm where the deconvolution map is permanently set according to the true bulk celltype percentages. Both dsLDA and NNLS (MuSiC) deconvolution approaches are included for DURIAN benchmarks. Two sets of values for the SCRABBLE objective are provided for DURIAN, SCRABBLE and mtSCRABBLE: 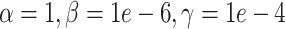 and
and 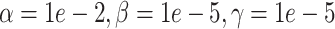 . (C) MA plots for a replicate of the downsampling strategy at sparsity
. (C) MA plots for a replicate of the downsampling strategy at sparsity 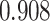 . The y-axis is the log ratio of true vs imputed gene-wise average. The x-axis is the log average over true and imputed gene-wise counts. (D) MA plots for a replicate of the simulation strategy at mean sparsity
. The y-axis is the log ratio of true vs imputed gene-wise average. The x-axis is the log average over true and imputed gene-wise counts. (D) MA plots for a replicate of the simulation strategy at mean sparsity 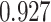 . DURIAN parameters for B–C are
. DURIAN parameters for B–C are  .
.
