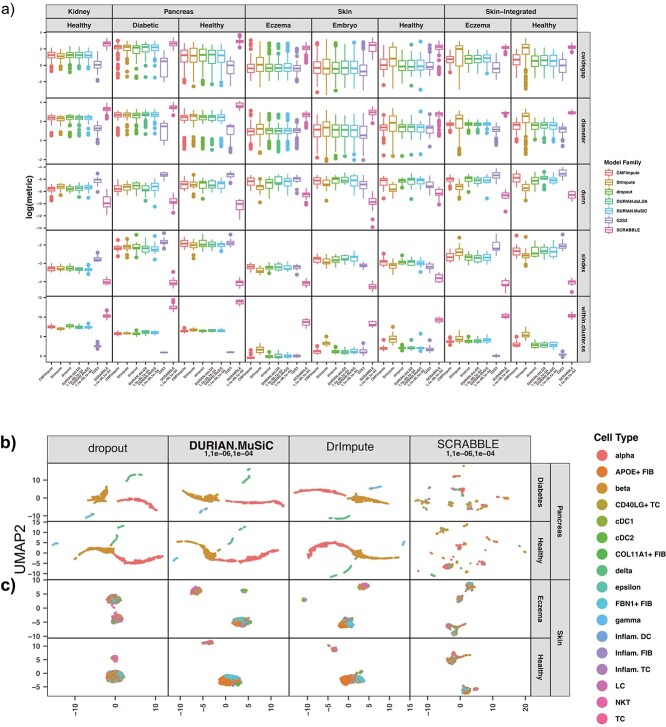
Figure 4.
Quantitation of cluster-quality for imputated single-cell data (A) Single-cell data from adult human pancreas (healthy and diabetic) and skin (healthy and eczema), and mouse embryonic skin was imputed via DrImpute, SCRABBLE and DURIAN. The y-axis corresponds to the log of the following cluster statistics: ‘cwidegap’ (lower is better): the longest edge in the within-cluster spanning tree of each cluster (each data point corresponds to a single cluster), ‘diameter’ (lower is better) the longest distance between two cells of a single cluster (each data point corresponds to a single cluster), ‘dunn’ (higher is better): each point represents the Dunn index corresponding to the minimum separation verus maximum diameter for two (possibly the same) clusters in the corresponding data set, ‘sindex’ (higher is better): the Silhouette Index corresponding to the mean of the bottom 10% of the distances from every cell in a cluster to the closest cell not in the same cluster (each data point corresponds to a single cluster), ‘within.cluster.ss’ (lower is better): half the sum of the squared distances for each pair of cells in the cluster divided by the cluster size (each data point corresponds to a single cluster). (B–C) Scatter plots of the imputed result from each data set, colored by the published label.