Abstract
COVID-19 has been spreading continuously since its outbreak, and the detection of its manifestations in the lung via chest computed tomography (CT) imaging is essential to investigate the diagnosis and prognosis of COVID-19 as an indispensable step. Automatic and accurate segmentation of infected lesions is highly required for fast and accurate diagnosis and further assessment of COVID-19 pneumonia. However, the two-dimensional methods generally neglect the intraslice context, while the three-dimensional methods usually have high GPU memory consumption and calculation cost. To address these limitations, we propose a two-stage hybrid UNet to automatically segment infected regions, which is evaluated on the multicenter data obtained from seven hospitals. Moreover, we train a 3D-ResNet for COVID-19 pneumonia screening. In segmentation tasks, the Dice coefficient reaches 97.23% for lung segmentation and 84.58% for lesion segmentation. In classification tasks, our model can identify COVID-19 pneumonia with an area under the receiver-operating characteristic curve value of 0.92, an accuracy of 92.44%, a sensitivity of 93.94%, and a specificity of 92.45%. In comparison with other state-of-the-art methods, the proposed approach could be implemented as an efficient assisting tool for radiologists in COVID-19 diagnosis from CT images.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11517-022-02619-8.
Keywords: COVID-19, Infected lesion segmentation, Screening, Computed tomography
Introduction
In December 2019, the novel coronavirus disease (COVID-19) broke out and rapidly developed into a global epidemic [1–7]. COVID-19 presents rapid spread, strong virulence, and high mortality in critically ill patients. To date, the disease has been continuously attacked 224 countries, areas, or territories involving 291,413,610 confirmed COVID-19 cases and 5,461,241 confirmed deaths according to World Health Organization (WHO). For COVID-19 screening, chest computed tomography (CT) imaging is considered as an indispensable tool for detection of manifestations in the lung associated with the disease [8–13]. The qualitative evaluation of infection in CT slices could provide important and useful information for COVID-19 diagnosis. However, the manual contouring of lung diseases requires tremendous time and effort. Automated tools can assist radiologists to accelerate autocontouring for COVID-19 infections in the clinical practice. Therefore, automated segmentation of COVID-19 from chest CT images is necessary for quantitative assessment.
Recently, several deep learning (DL) methods have been developed for segmentation of COVID-19 in CT images [6, 14–22]. Serena et al. [23] reviewed the application of deep learning techniques in COVID-19 chest of pneumonia cases. Li et al. [23] employed the UNet to segment the lungs from CT scans to distinguish COVID-19 pneumonia from community acquired pneumonia. Fan et al. [24] presented a COVID-19 lung infection segmentation deep network (Inf-Net) for CT images and introduced a semi-supervised segmentation system for COVID-19 lung infection. Gao et al. [25] developed a dual-branch combination network (DCN) for COVID-19 diagnosis that can simultaneously achieve individual-level classification and lesion segmentation. Moreover, Zhou et al. [21] proposed a fully automatic, rapid, accurate, and machine-agnostic method that can segment and quantify the infection regions on CT scans from different sources, which decomposes the 3D segmentation problem into three 2D ones, thereby reducing the model complexity by an order of magnitude. Shan et al. [17] proposed a 3D segmentation method referred to as VB-Net. V-Net was combined with a bottleneck structure [26] to segment multiple structures including lungs, lung lobes, and infection regions.
It has to note that automatic and accurate segmentation of the infection region is still very challenging owing to the high variation in size, shape, and distribution of lesions in 3D images. Moreover, some lesions, i.e., ground-glass opacity (GGO), have low contrast and irregularity in boundaries, leading to a degradation of the performance of solely interslice-based segmentation methods. Hence, we propose a DL-based automatic segmentation network for the quantitative assessment of infected lesions for COVID-19, based on the hybrid densely connected UNet proposed by Li et al. [27] and the skip pathways proposed by Zhou et al. [28]. The proposed network consists of two parts, a 2D network and a 3D network, referred to as two-stage hybrid UNet (TSH-UNet). The deep and efficient 2D network inherits the structure of UNet which is used to extract intraslice features and obtain a rough segmentation result of the CT images. In addition, compared with the method proposed by Li et al. [27], our model adopts long-range and short-range skip connections between the encoding part and the decoding part to enable low-level spatial feature preservation for better intraslice context exploration. The 3D network has a structure similar to that of the 2D network. Compared with the method proposed by Zhou et al. [28], it takes the context on the intraslice direction into consideration to effectively extract intraslice features and 3D contexts, which are jointly optimized for accurate lung and infected lesion segmentation. We demonstrate the superiority of the proposed method over state-of-the-art methods from multiple perspectives. The main contributions of this study are summarized as follows:
We propose a novel lung and infected lesion segmentation network, TSH-UNet, which can extract the features of different layers and fully use context information, thereby balancing segmentation accuracy and network complexity.
We verify the performance of our network on datasets obtained from multiple centers to show the generalization capabilities of our network, which can be applied to other centers in the future.
We use the results of the segmentation network to extract the lesions from the original CT images to classify COVID-19 pneumonia and other common types of pneumonia in an existing classification model (3D ResNet) [29], and the results are superior to those obtained through direct classification with the original image.
Methods
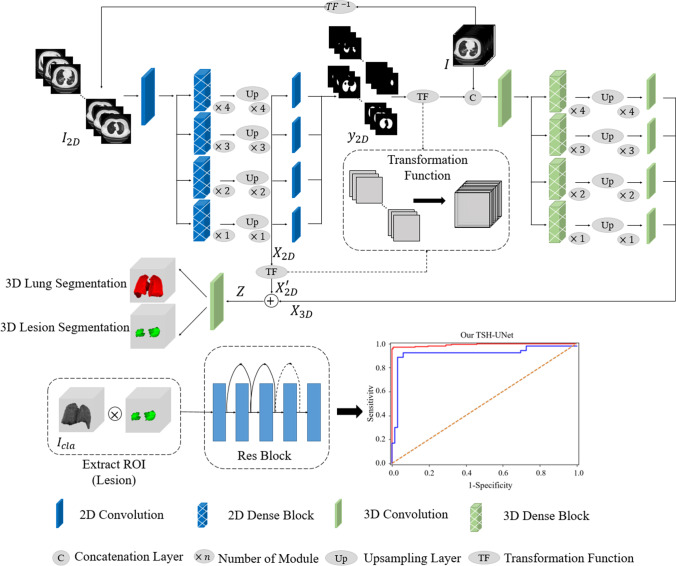
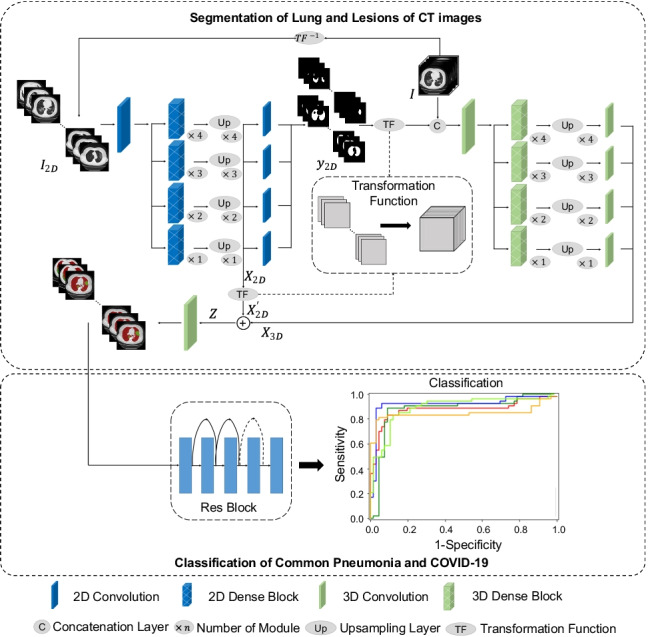
Herein, we developed a model for segmentation and classification tasks based on DL network for COVID-19 screening, as shown in Fig. 1. To achieve accurate lung and lesion segmentation, our segmentation network was constructed using a TSH-UNet combining a 2D network and a 3D network. First, we trained the 2D network to obtain a coarse segmentation result; specifically, the 2D network adopts the skip pathway from UNet + + [28, 30], where representative high-quality intraslice features can be extracted with deep convolutions. Then, a fine-tuning procedure was performed by jointly optimizing the 2D and 3D networks, where the interslice and intraslice features can be fully explored. For the classification task, the patients with COVID-19 pneumonia and other pneumonia were distinguished using 3D ResNet [29] with the segmentation results.
Fig. 1.
Main framework of the proposed model. The structure of TSH-UNet for lung and lesion segmentation includes a 2D network and a 3D network, the detailed architecture of which can be found in supplementary Fig. 1 and supplementary Table 1. The input volume data are transformed into three consecutive slices and fed into the 2D network, yielding a coarse segmentation result. Then, after concatenation with the predicted volumes from 2D network, the input volume data are fed into 3D network to extract interslice features. Finally, the hybrid features are jointly optimized in the 3D network to accurately segment lungs and lesions. For classification, the region of interest is extracted using the segmentation result. The 3D network used in the classification part of our model is adapted from 3D ResNet. Finally, the model outputs the probability of COVID-19 pneumonia and common pneumonia. The 2D network consisted of 167 layers, including convolutional layers, pooling layers, upsampling layers, transition layers, and a dense block. The dense block represented the cascade of several microblocks, where all layers were directly connected. The transition layers were used to resize the feature maps, which were composed of a batch normalization layer and convolutional layer (1 × 1) followed by an average pooling layer. We set the compression factor as 0.5 in the transition layers to prevent the feature maps from expanding. Bilinear interpolation was employed in the upsampling layer, followed by the sum of low-level features and a convolutional layer (3 × 3). In addition, batch normalization and the rectified linear unit were used before each convolutional layer in the architecture
Segmentation network
Training 2D network
We used the 2D network in the first feature extraction stage to obtain a preliminary result. The UNet structure with long-range and short-range skip connections was used to extract the features of different levels of COVID-19 CT images. The skip pathways change the connectivity of encoder and decoder subnetworks to aggregate the features of varying semantic scales, leading to a flexible feature fusion scheme.
We set as the training samples with ground-truth labels . because each pixel was identified as class , where could be the lung, lesion, or background. Let function denote the transformation from three adjacent slices to volumetric data. denotes the inverse transformation from volumetric data to three adjacent slices. We transformed the input training samples, , from volumetric data to three slices to obtain the input of 2D network, (see Fig. 1). The 2D network segment lungs and lesions as follows:
| 1 |
| 2 |
where denotes the feature map from the “upsampling layer,” and denotes the corresponding pixel-wise probabilities for input . represents the partial 2D network before the upsampling layer, and represents the last layer of the 2D network. and denote the parameters in the corresponding network.
The 2D network consisted of 167 layers, including convolution layers, pooling layers, upsampling layers, transition layers, and a dense block. The dense block represented the cascade of several microblocks, where all layers were directly connected. The transition layers were used to resize the feature maps, which were composed of a batch normalization layer and convolution layer (1 1) followed by an average pooling layer. We set the compression factor as 0.5 in the transition layers to prevent the feature maps from expanding. Bilinear interpolation was employed in the upsampling layer, followed by the sum of low-level features and a convolutional layer (3 3). In addition, batch normalization and the rectified linear unit were used before each convolution layer in the architecture.
Training 2D and 3D network
The 2D network can accurately extract intraslice features but neglects the spatial information between slices, which would lead to rough results. Compared to the 2D network, the 3D network can learn spatial information, but it is limited in terms of the field of view of the kernel and depth of the network, in addition to incurring a high computational cost. To address these issues, we propose the two-stage hybrid network to jointly fuse and optimize the learned intraslice and interslice features for better lung and lesion segmentation.
First, we obtained a preliminary segmentation result from the 2D network. Subsequently, we set the transformation function (see Fig. 1) to obtain volumetric data from the slices to fine-tune the model. Thus, the feature map, , and output of 2D network, , were transformed from slices to volumetric data, and . Then, we concatenated the original samples, with the contextual output information, , and fed it into the 3D network to extract visual features with 3D contexts. In this study, the 3D network was composed of 65 convolutional layers, with a growth rate of 32. In comparison with its 2D counterpart, the 3D network decreased the number of microblocks in each dense block, resulting in low GPU memory consumption and high memory consumption for 3D convolutions. The other part of the 3D network was similar to that of the 2D counterpart. In this manner, the 3D network was trained based not only on the features distilled from original images but also on the pre-defined coarse segmentation results from the 2D network. The guidance from supporting context pixels made it easier to find the optimal solution, which improved the efficiency of the 3D network. The training process of 3D network is as follows:
| 3 |
| 4 |
where is the volumetric feature extracted from 3D network, and represents the hybrid feature, which is the sum of intraslice and interslice features. represents the 3D network, and represents the parameters thereof. Then, the hybrid feature is trained:
| 5 |
where denotes the predicted pixel-wise probabilities obtained from , and denotes the parameters in the corresponding network. The architecture information of 2D and 3D networks is detailed in the supplementary Table 1. The input CT slice/volume type is Hounsfield units (HU).
Loss function and training strategy
During training, the weight cross-entropy function [31] was employed as the loss function, which is expressed as follows:
| 6 |
where represents the possibility that voxel belongs to class , represents the label of voxel in the ground truth, and denotes the weight.The logarithm base is 2, the weights of the loss function are as follows: 0.78, 0.65, and 8.57.
To train TSH-UNet, we employed transfer learning [32–34] to improve the performance of the network. First, the parameters of the encoder part in the 2D network were obtained from pre-trained DenseNet (object classification trained) [35] while the decoder part are trained with the random initialization. We first trained the 2D network separately to extract intraslice features and obtain a rough segmentation result. The 3D network was fine-tuned with the result produced by the 2D network, concatenating with the original 3D chest CT volume. Finally, we jointly optimized the 3D interslice features along with the 2D intraslice features to yield accurate segmentation of lung and lesion.
Python version 3.6 was used in our deep learning model, which was implemented in the open framework tensorflow (version: 1.4.0) and keras (version: 2.0.8). The model was run on a single NVIDIA Titan GPU. TSH-UNet was trained for 500 epochs using the SGD optimizer with a batch size of 1. The initial learning rate was set as 0.01, and it decayed according to the equation , where denotes the iterations step, and denotes the total iteration number. For data augmentation, different transformations, including scaling between 0.8 and 1.2 and random mirroring, were randomly applied to all training data to prevent the overfitting problem. After data enhancement, the total number of slices during training was 24,192. The test data were input into the trained TSH-UNet, and the network automatically segmented the lung and lesions of COVID-19.
Classification network
We trained a classification network for diagnostic analysis after our segmentation network operation to distinguish COVID-19 pneumonia from other types of pneumonia (see Fig. 1). The input of the diagnostic classification model was the intersection of raw image and the segmentation results, and further diagnostic prediction was made using CT volumetric data with and without COVID-19 pneumonia. As TSH-UNet was trained using CT volumetric data, the network used in the classification part of our model was also a 3D classification model. We normalized the height, width, and depth of the segmentation results to 128 × 128 × 64. The 3D classification network considered a tensor as the input and output the diagnostic probability of COVID-19 pneumonia and other common types of pneumonia. The 3D network used in the classification part of our model was adapted from 3D ResNet [29], which has five layers. The first layer is a normal 1 1 1 convolutional layer and the second to fourth layers are residual layers. Each layer is composed of three residual modules. The multiple residual connections were used to continuously extract local and global context features. A fully connected layer and soft-max activation function were used to calculate the predicted diagnostic probability. Finally, the output of the network was the category of the maximum probability. In addition, the cross-entropy loss between the predicted results and ground-truth labels was used to train the 3D classification network. We used the stochastic gradient descent (SGD) optimizer to train the network, and the training batch size was 4.
Evaluation metrics
The Dice coefficient and Hausdorff distance were employed to evaluate the lung and lesion segmentation performance. The Dice coefficient [36] is typically used to calculate the similarity or overlap between two samples, and it represents the degree of coincidence between the segmentation result and ground-truth labels. The Hausdorff distance [37] indicates the similarity between two sets. In addition, we applied four other metrics [38] to evaluate the accuracy of the segmentation results, i.e., the volumetric overlap error (VOE), relative volume difference (RVD), average symmetric surface distance (ASD), and root mean square symmetric surface distance (RMSD). Moreover, we compared the segmentation results with the ground-truth labels to quantify the accuracy value of the results.
The confusion matrix was employed to evaluate the performance of the classification. From the confusion matrix, false positive (FP), false negative (FN), true positive (TP), and true negative (TN) values were obtained, four metrics for performance evaluation were calculated as below: accuracy, sensitivity, specificity, and area under the receiver-operating characteristics curve (AUC).
Datasets
Dataset for segmentation
This study was approved by the multiple institutional review board of Renmin Hospital of Wuhan University (RHWU), Anhui Medical University (AMU), Beijing Chaoyang Hospital (BCH), Beijing Youan Hospital (BYH), Zhuhai Renmin Hospital (ZRH), Henan Provincial Renmin Hospital (HPRH), and Hubei Huangshi Central Hospital (HHCH), all in China. Written informed consent was waived by the institutional review board for the retrospective study.
We collected high-resolution CT images of 103 patients with COVID-19 pneumonia from the abovementioned hospitals in China, including 48 cases of COVID-19 from RHWU (Optima CT680, GE Medical Systems; tube voltage: 140 kVp; slice thickness: 0.625 mm), 14 cases from BYH (iCT 256, Philips; tube voltage: 140 kVp; slice thickness: 0.9 mm), 9 cases from ZRH (SOMATOM Definition Flash, Siemens; tube voltage: 120 kVp; slice thickness: 10 mm), and 8 cases each from AMU (LightSpeed VCT, GE Medical Systems; tube voltage: 120 kVp; slice thickness: 5 mm), BCH (NeuViz 64 In, NMS; tube voltage: 120 kVp; slice thickness: 1 mm), HPRH (CT 780, UIH; tube voltage, 120 kVp; slice thickness: 1 mm), and HHCH (Perspective, Siemens; tube voltage: 130 kVp; slice thickness: 7 mm). We divided the data from RHWU into a training dataset (28 cases), validation dataset (8 cases), and testing dataset (12 cases) and set the dataset from other centers as the external testing dataset. The dataset and demographic characteristics of patients for segmentation tasks are provided in Table 1.
Table 1.
Datasets and demographic characteristics of patients with COVID-19 pneumonia for segmentation tasks
| Cohorts | Cases | Slices | Gender | Age | |
|---|---|---|---|---|---|
| M | F | ||||
| Training dataset (n = 28) | 28 | 8,064 | 15 | 13 | 56 ±19 (24–86) |
| Validation dataset (n = 8) | 8 | 2,160 | 1 | 7 | 51 ±14 (25–68) |
| Testing dataset (n = 12) | 12 | 5,617 | 7 | 5 | 61 ±14 (30–85) |
| Total | 48 | 15,841 | 23 | 25 | 55 ±17 (24–86) |
| Independent cohorts (external testing dataset) | |||||
| AMU (n = 8) | 8 | 235 | 3 | 5 | 50 ±17 (21–86) |
| BCH (n = 8) | 8 | 666 | 4 | 4 | 43 ±21 (24–92) |
| BYH (n = 14) | 14 | 1,761 | 6 | 8 | 43 ±14 (17–68) |
| ZRH (n = 9) | 9 | 583 | 5 | 4 | 51 ±20 (25–80) |
| HPRH (n = 8) | 8 | 2,528 | 5 | 3 | 43 ±13 (22–63) |
| HHCH (n = 8) | 8 | 344 | 4 | 4 | 53 ±19 (27–86) |
| Total | 55 | 6,117 | 27 | 28 | 47 ±17 (17–92) |
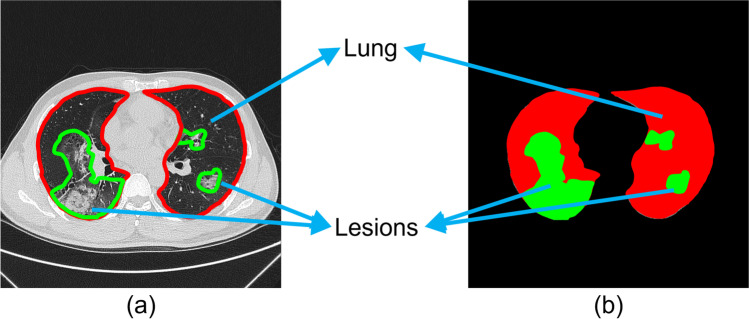
The lesions were manually annotated by trained radiologists from RHWU, AMU, BCH, BYH, ZRH, HPRH, and HHCH as the ground-truth labels. The region of the lung was first delineated slice-by-slice. As the shapes of the lesions were generally irregular, we marked all representative lesions and suspicious lesions. All the radiologists were asked to jointly evaluate suspicious lesions to reduce human error. We annotated the ground-truth labels as background (black), lung (red), and lesions (green), as shown in Fig. 2.
Fig. 2.
a Raw CT image: the red and green lines represent the outlines of lung and lesions, respectively. b Ground-truth label: the black, red, and green regions represent background, lung, and lesions, respectively.
In addition, we chose the online datasets (COVID-19 CT Lung and Infection Segmentation Dataset) [39]. This dataset contains raw and labeled COVID-19 CT scans. Lung and infections are labeled by two radiologists and verified by an experienced radiologist.
Dataset for classification
The dataset of patients for classification tasks are provided in Table 2. We collected the high-resolution CT images of 439 patients from RHWU, including 224 patients with clinical confirmed COVID-19 pneumonia and 215 patients with other common types of pneumonia, which was divided into a training dataset (160 COVID-19, 160 other pneumonia) and test dataset (64 COVID-19, 55 other pneumonia). We also collected data on 184 cases from AMU (30 COVID-19, 33 other pneumonia; Aquilion, Toshiba; tube voltage: 120 kVp; slice thickness: 5 mm), BYH (30 COVID-19, 31 other pneumonia; iCT 256, Philips; tube voltage: 120 kVp; slice thickness: 5 mm), and ZRH (30 COVID-19, 30 other pneumonia; SOMATOM Definition Flash, Siemens; tube voltage: 120 kVp; slice thickness: 5 mm) for an external validation dataset.
Table 2.
Datasets and demographic characteristics of patients with COVID-19 pneumonia for classification tasks
| Cohorts | COVID-19 | Other pneumonia | Total |
|---|---|---|---|
| Training dataset (n = 320) | 160 | 160 | 320 |
|
Testing dataset (n = 119) |
64 | 55 | 119 |
| Total | 224 | 215 | 439 |
| Independent cohorts (external validation dataset) | |||
| AMU (n = 63) | 30 | 33 | 63 |
| BYH (n = 61) | 30 | 31 | 61 |
| ZRH (n = 60) | 30 | 30 | 60 |
| Total | 90 | 94 | 184 |
For segmentation and classification datasets, we converted all CT images from the DICOM format into the 3D NII format to train our model. The threshold value of the CT images was normalized to the range of [− 1000, 200]. The last step was to find the largest connected domain in the image to distinguish the background from the lung. This step was to extract region of interest and remove unnecessary background.
Results
Segmentation results
To analyze the performance of our TSH-UNet on a volumetric dataset, we compared it with several semantic segmentation networks, including the fully convolutional network (FCN) [40], SegNet [41], Li et al. [23], Wang et al. [42], Müller et al. [43], and H-DenseUNet [27]. Li et al. [23] was using 2D UNet. Wang et al. [42] was using 2D UNet + + , Müller et al. [43] was using 3D UNet + + . We tested our TSH-UNet on the testing dataset from RHWU, and the evaluation metrics were calculated as listed in Table 3. As for the metric of lung, TSH-UNet achieved a dice of 97.00%. For the metric of lesions, TSH-UNet achieved a dice of 89.22%, outperforming other networks. The Dice and accuracy of lung and lesion segmentation using TSH-UNet was the highest. The Dice of the lung, Dice of the lesion, accuracy of the lung, and accuracy of lesion were 0.62–8.88%, 0.57–16.22%, 0.80–4.08%, and 1.17–20.10% higher than those of other networks, respectively.
Table 3.
Comparison of lung and lesion segmentation results on the test dataset
| Model | Lung | ||||||
| Dice (%) | Hausdorff (mm) | VOE (%) | RVD (%) | ASD (mm) | RMSD (mm) | Accuracy (%) | |
| FCN | 88.12 ±3.18 | 42.47 ±27.30 | − 9.61 ±16.97 | − 7.81 ±16.85 | 18.53 ±6.87 | 1.35 ±0.61 | 93.15 ±8.78 |
| SegNet | 95.47 ± 2.84 | 37.59 ±47.08 | − 5.16 ±6.76 | − 4.83 ±6.29 | 13.53 ±10.58 | 0.91 ±0.56 | 96.43 ±1.35 |
| Li et al. [23] | 95.85 ± 2.11 | 20.30 ±7.45 | 3.39 ±6.46 | − 3.98 ±5.77 | 8.55 ±6.76 | 0.63 ±0.47 | 95.38 ±2.03 |
| Wang et al. [42] | 95.72 ± 2.26 | 20.55 ±8.11 | − 5.15 ±3.82 | − 4.85 ±3.55 | 9.61 ±7.98 | 0.79 ±0.60 | 96.06 ±0.35 |
| Müller et al. [43] | 95.96 ±1.92 | 10.09 ±2.74 | − 5.07 ±6.47 | − 4.75 ±6.29 | 4.75 ±6.28 | 0.34 ±0.35 | 95.15 ±2.36 |
| H-DenseUNet | 96.38 ±1.64 | 11.14 ±3.15 | 2.15 ±1.12 | 2.19 ±1.16 | 3.48 ±2.26 | 0.32 ±0.26 | 96.63 ±0.03 |
| TSH-UNet | 97.00 ±1.26 | 8.95 ±2.23 | − 0.44 ±2.44 | − 0.41 ±2.41 | 2.73 ±2.10 | 0.27 ±0.28 | 97.23 ±0.93 |
| Model | Lesion | ||||||
| Dice (%) | Hausdorff (mm) | VOE (%) | RVD (%) | ASD (mm) | RMSD (mm) | Accuracy (%) | |
| FCN | 73.00 ±7.86 | 23.67 ±8.64 | 29.71 ±25.11 | 40.14 ±36.03 | 11.25 ±0.60 | 0.71 ±0.40 | 64.48 ±12.52 |
| SegNet | 81.98 ±5.65 | 17.69 ±5.02 | 10.62 ±16.34 | 12.91 ±19.72 | 8.54 ±5.09 | 0.70 ±0.49 | 78.44 ±9.09 |
| Li et al. [23] | 84.88 ±2.55 | 17.90 ±6.69 | 10.71 ±10.95 | 12.04 ±12.52 | 7.92 ±4.26 | 0.63 ±0.38 | 80.84 ±5.74 |
| Wang et al. [42] | 84.44 ±2.75 | 18.28 ±5.89 | 13.19 ±9.57 | 14.71 ±11.56 | 7.19 ±4.19 | 0.58 ±0.39 | 79.44 ±5.35 |
| Müller et al. [43] | 88.65 ±3.25 | 10.54 ±2.72 | − 6.16 ±8.71 | − 5.62 ±8.32 | 3.97 ±1.56 | 0.31 ±0.18 | 83.41 ±5.17 |
| H-DenseUNet | 83.80 ±2.88 | 15.70 ±5.11 | 16.56 ±13.13 | 19.15 ±15.30 | 6.66 ±2.85 | 0.49 ±0.26 | 77.81 ±7.01 |
| TSH-UNet | 89.22 ±1.93 | 8.47 ±2.55 | 12.24 ±8.18 | 7.33 ±3.63 | 3.64 ±1.52 | 0.29 ±0.15 | 84.58 ±3.48 |
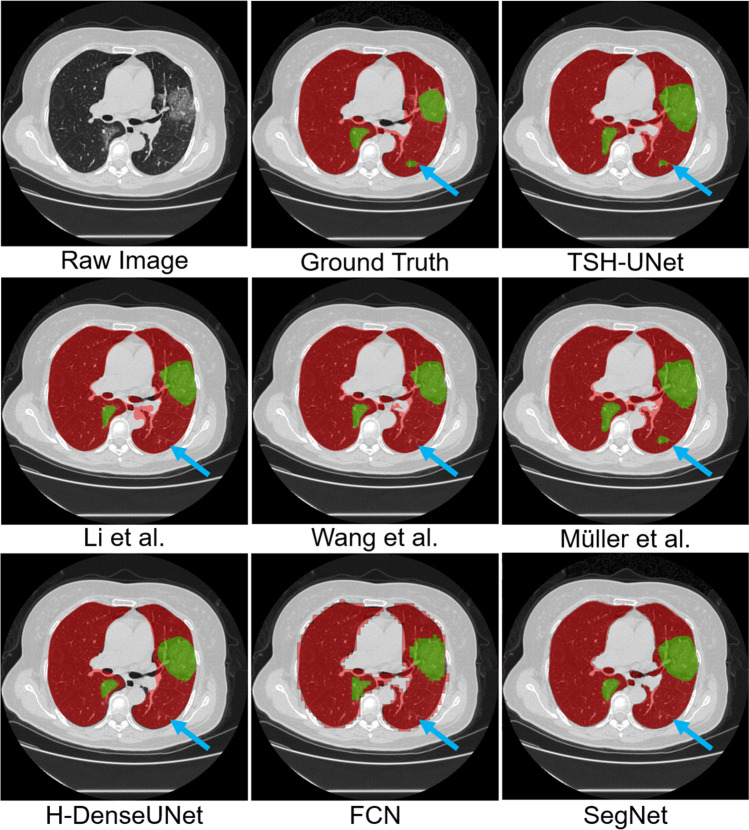
The representative segmentation results on the testing dataset of different networks are shown in Fig. 3. It is clear that TSH-UNet delivered the best performance in terms of both lung and lesions. The FCN segmented the contour of the lung, but the edges were zigzag-shaped, which caused errors. SegNet and Li et al. [23] obtained relatively accurate results for lung segmentation. However, their performance was poorer that of TSH-UNet in terms of the Dice coefficient and accuracy. Moreover, the segmentation result of our TSH-UNet was better than that of H-DenseUNet in lung and lesion, and we attribute this improvement to long-range and short-range skip connections.
Fig. 3.
Representative segmentation results of testing dataset obtained from different segmentation networks. We selected one typical slice as an example to elucidate the difference between results
From Fig. 3, it is observed that TSH-UNet can achieve better results than the 2D network. Our TSH-UNet can accurately segment small lesions (see the blue arrows in Fig. 3) that the 2D network cannot, which explains the second stage hybrid architecture of our model, a fine-tuning procedure that obtains a more accurate result and improves the dice and accuracy of both lung segmentation and lesion segmentation. The trained TSH-UNet operates in an end-to-end manner, where the 3D contexts can also help extract more representative interslice features. The two-stage hybrid architecture jointly optimizes the 2D and 3D networks, where the hybrid feature can be fully explored. With the hybrid features extracted from the 2D slices and 3D contexts on the z-axis, our model can segment small lesions, which the 2D network neglects (see the blue arrows in Fig. 3). The 3D display of the results of Wang et al. [42] and our TSH-UNet can be found in supplementary Fig. 2. Moreover, we found that only Müller et al. [43] can achieve better performance than that of the Wang et al. [42]. Because of the extraction of the 3D contents along the intraslice direction. However, 3D kernel consumes large GPU memory, because of which Müller et al. [43] took much more training time to converge compared to that required for Wang et al. [42]. The training of Wang et al. [42] took about 12 h using a single NVIDIA Titan GPU, while two-stage hybrid fine-tuning took 5 h. In other words, the total training time for TSH-UNet was about 17 h, while that of Müller et al. [43] was around 35 h.
We validate our method and comparison methods on the online datasets to prove the generalization performance of our TSH-UNet. The experimental results are summarized in Table 4 and Fig. 4. Compared to the results in Table 3, the results of the online datasets were slightly worse, but our method achieved reasonable performance on the results of both lung segmentation and lesion segmentation. For lung segmentation, the Dice coefficients of different methods were higher than 80%, and the accuracy were above 80%. For lesion segmentation, the Dice coefficients of TSH-UNet was 67.23%, and the accuracy was 73.44%.
Table 4.
Comparison of lung and lesion segmentation results on the online datasets
| Model | Lung | |||
| Dice (%) | VOE (%) | RVD (%) | Accuracy (%) | |
| FCN | 82.58 ±4.64 | 4.37 ±2.71 | 4.24 ±2.59 | 84.44 ±5.05 |
| SegNet | 82.20 ±4.29 | 12.66 ±3.81 | 11.85 ±3.32 | 87.83 ±5.43 |
| Li et al. [23] | 84.38 ±4.53 | 6.38 ±2.03 | 6.16 ±1.92 | 87.19 ±5.13 |
| Wang et al. [42] | 84.38 ±4.53 | 6.38 ±2.03 | 6.16 ±1.92 | 87.19 ±5.13 |
| Müller et al. [43] | 81.29 ±4.86 | 4.51 ±2.77 | 2.59 ±4.42 | 82.49 ±5.94 |
| H-DenseUNet | 83.86 ±4.60 | 6.82 ±5.95 | 7.10 ±6.59 | 81.25 ±5.50 |
| TSH-UNet | 85.49 ±3.98 | 1.95 ±1.19 | 0.13 ±2.26 | 85.59 ±4.48 |
| Model | Lesion | |||
| Dice (%) | VOE (%) | RVD (%) | Accuracy (%) | |
| FCN | 63.97 ±3.84 | 21.31 ±11.92 | 7.99 ±20.86 | 68.43 ±8.38 |
| SegNet | 64.90 ±2.58 | 33.77 ±17.77 | 5.13 ±36.19 | 72.03 ±15.76 |
| Li et al. [23] | 62.48 ±4.14 | 27.93 ±16.09 | 9.05 ±25.71 | 68.62 ±11.27 |
| Wang et al. [42] | 65.55 ±2.48 | 25.86 ±18.67 | 14.32 ±22.32 | 73.89 ±11.78 |
| Müller et al. [43] | 56.98 ±4.83 | 72.96 ±16.38 | 52.92 ±8.93 | 90.29 ±5.30 |
| H-DenseUNet | 55.03 ±3.94 | 40.84 ±19.16 | 32.86 ±13.26 | 70.32 ±10.84 |
| TSH-UNet | 67.23 ±0.65 | 23.39 ±7.95 | 12.17 ±18.59 | 73.44 ±7.56 |
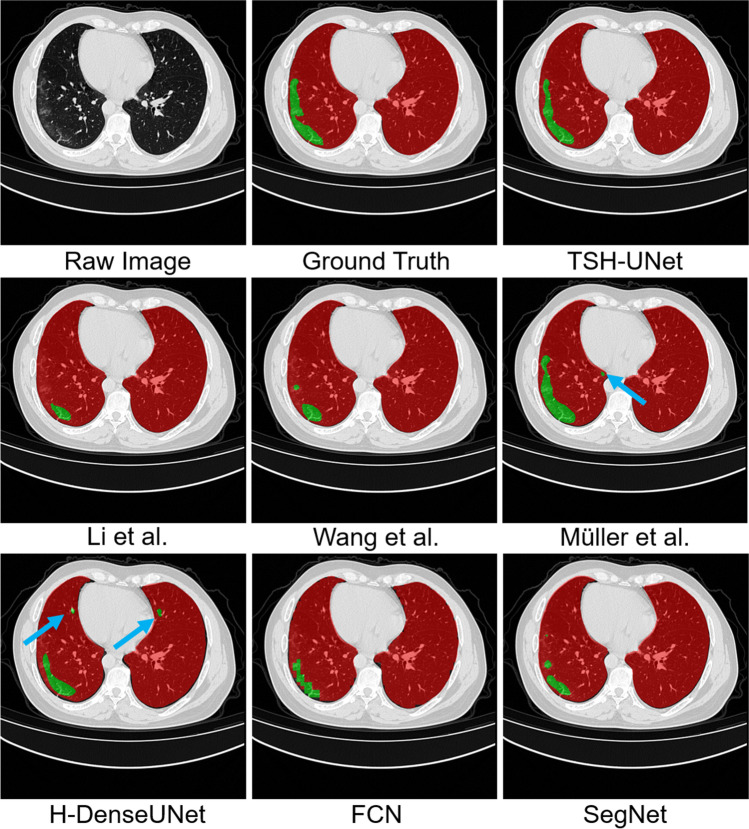
Fig. 4.
Representative segmentation results of the online datasets obtained from different segmentation networks. We selected one typical slice as an example to elucidate the difference between results
From Fig. 4, we can find that the three 3D networks, Müller et al. [43], H-DenseUNet, and our TSH-UNet can better segment the regions of lesion because of the extraction of the 3D contents along the intraslice direction. However, some small regions of lesion were segmented in the results of Müller et al. [43] and H-DenseUNet (see the blue arrows). Our TSH-UNet can accurately segment lesions because of the second-stage hybrid architecture of our model, fine-tuning our model to obtain a more accurate result.
Multicenter verification for segmentation tasks
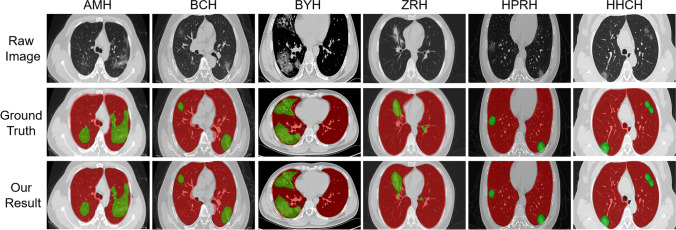
We randomly selected 55 cases from six hospitals to illustrate the effectiveness and generalization performance of our TSH-UNet. We selected 8 cases each from AMU, BCH, HPRH, and HHCH, 9 cases from ZRH, and 14 cases from BYH as the external validation dataset to verify our model. A few representative metrics are shown in Table 5, and the results are shown in Fig. 5. The segmentation of the lung and different lesions was satisfactory. For lung segmentation, the Dice coefficients of all centers were 94%, and the accuracy was above 97%. For lesion segmentation, the Dice coefficients were approximately 80%, and the highest accuracy was 88.69%.
Table 5.
Representative evaluation metrics of the segmentation results for different centers
| Lung | ||||
| Dice (%) | VOE (%) | RVD (%) | Accuracy (%) | |
| AMU | 97.25 | 5.18 0.76 | 5.04 0.72 | 99.80 1.20 |
| BCH | 98.40 0.41 | 2.53 0.87 | 2.49 0.85 | 99.66 0.40 |
| BYH | 98.55 0.21 | 2.72 0.48 | 2.68 0.46 | 99.91 0.21 |
| ZRH | 98.26 0.77 | 3.14 1.50 | 3.08 1.43 | 99.83 0.13 |
| HPRH | 94.91 1.18 | 6.77 1.94 | 6.53 1.81 | 98.25 1.54 |
| HHCH | 94.62 1.16 | 5.81 3.27 | 5.60 3.12 | 97.47 1.27 |
| Lesion | ||||
| Dice (%) | VOE (%) | RVD (%) | Accuracy (%) | |
| AMU | 78.49 4.58 | 20.06 9.14 | 17.92 7.59 | 87.48 6.75 |
| BCH | 79.06 6.32 | 12.86 5.37 | 11.97 4.82 | 84.29 7.83 |
| BYH | 82.38 5.64 | 16.30 10.41 | 14.74 8.94 | 88.69 7.60 |
| ZRH | 78.36 5.20 | 16.78 8.07 | 18.73 9.52 | 72.78 7.50 |
| HPRH | 79.22 3.27 | 17.02 9.49 | 15.34 7.88 | 86.71 3.63 |
| HHCH | 77.77 3.17 | 16.93 6.43 | 17.98 8.12 | 78.15 8.08 |
Fig. 5.
Representative segmentation results of different centers. We selected one typical slice from the datasets as an example
Classification results
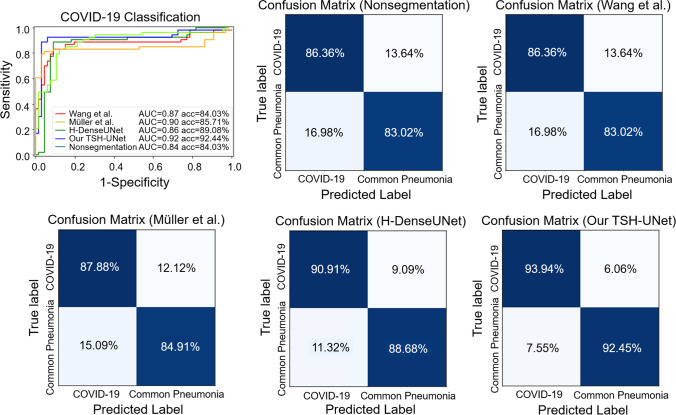
To investigate the performance of COVID-19 classification tasks with and without segmentation, we trained a 3D ResNet to identify COVID-19 pneumonia using the intersection of raw image and the lesion segmentation results as the input. With the lesion segmentation results of our network, the classification network could achieve a more accurate result in test datasets, as shown in Fig. 6. The classification network achieved an accuracy of 84.03%, a sensitivity of 86.36%, a specificity of 83.02%, and an AUC of 0.84 for COVID-19 screening without using segmentation results. When the segmentation results of TSH-UNet were used, the network could distinguish COVID-19 from other common types of pneumonia with an accuracy of 92.44%, a sensitivity of 93.94%, a specificity of 92.45%, and an AUC of 0.92. The experimental results showed that the performance of could be improved using segmentation results.
Fig. 6.
ROC curves and confusion matrixes of the classification model using results of different segmentation network on the testing datasets
We also compared the classification results by using the lesion segmentation results that outputted from Wang et al. [42], Müller et al. [43], and H-DenseUNet, respectively. For Wang et al. [42], the classification network could distinguish COVID-19 from other common types of pneumonia with an accuracy of 84.03%, a sensitivity of 86.36%, a specificity of 83.02%, and an AUC of 0.87. While for Müller et al. [43], the network achieved an accuracy of 85.71%, a sensitivity of 87.88%, a specificity of 84.91%, and an AUC of 0.90. Accordingly, the network achieved an accuracy of 89.08%, a sensitivity of 90.91%, a specificity of 88.68%, and an AUC of 0.86 with the results of H-DenseUNet. The similar results were also achieved on training datasets (see Supplementary Fig. 3 and Supplementary Fig. 4).
Multicenter verification for classification tasks
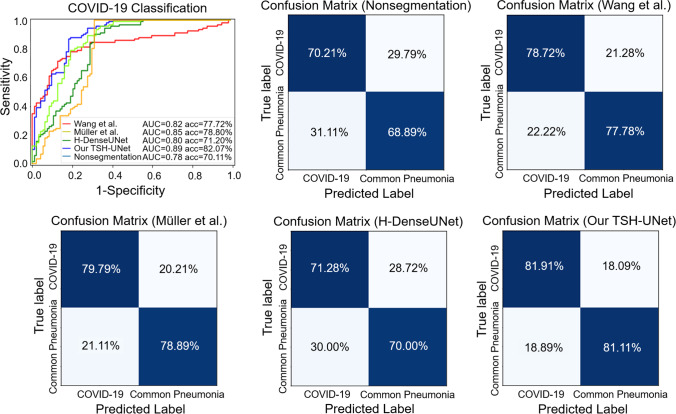
Our classification network was validated using multicenter data to demonstrate its generalization performance. We chose 63 cases from BYH (including 30 patients with COVID-19 and 33 patients with other common types of pneumonia), 61 cases AMU (including 30 patients with COVID-19 and 31 patients with other common types of pneumonia), and 60 cases from ZRH (including 30 patients with COVID-19 and 30 patients with other common types of pneumonia) as the test datasets to verify TSH-UNet. We directly fed the multicenter data into the trained model to obtain multicenter diagnosis results.
As shown in Fig. 7, the network achieved an accuracy of 70.11%, a sensitivity of 70.21%, a specificity of 68.89%, and an AUC of 0.78 without the input of segmentation results. When the segmentation results of TSH-UNet were used, the network could distinguish COVID-19 from other common types of pneumonia with an accuracy of 82.07%, a sensitivity of 81.91%, a specificity of 81.11%, and an AUC of 0.89. The network achieved an accuracy of 77.72%, a sensitivity of 78.72%, a specificity of 77.78%, and an AUC of 0.82 with the results of Wang et al. [42]. When the segmentation results of Müller et al. [43]were used, the network could distinguish COVID-19 from other common types of pneumonia with an accuracy of 78.80%, a sensitivity of 79.79%, a specificity of 78.89%, and an AUC of 0.85. The network achieved an accuracy of 71.20%, a sensitivity of 71.2%, a specificity of 70.00%, and an AUC of 0.80 with the results of H-DenseUNet.
Fig. 7.
Classification performance of the model on datasets obtained from AMU, BYH, and ZRH
Discussion
DL-based techniques could provide accurate and efficient solutions in COVID-19 diagnosis, prognosis, and treatment. Automatic lung and lesion segmentation of lungs, lobes, and infected regions or lesions in chest CT images could assist physicians in the clinical COVID-19 diagnosis process and further in quantification and assessment.
We proposed a DL-based two-stage network for quantitative infected lesion assessment for COVID-19. The main structure of our framework, i.e., TSH-UNet, was a combination of a 2D network (UNet structure) and a 3D network (UNet structure). Our TSH-UNet effectively extracted intraslice features and 3D contexts in intraslice direction, which were used for accurate lung and infected lesion segmentation. We employed an efficient and deep 2D network to extract sufficient intraslice features. The advantages of long-range and short-range skip connections and a densely connected path were combined in our 2D network. Through combination of the rough result output by the 2D network and the raw images, the 3D network was fine-tuned. Finally, the accurate segmentation was obtained by jointly optimizing the 2D intraslice features and 3D interslice features in the 3D network. The results showed that the performance of TSH-UNet was better than that of state-of-the-art networks for medical image segmentation.
In addition, we verified the generalization performance of our TSH-UNet on CT image datasets obtained from different centers. The CT protocols and settings of CT images, including CT equipment and slice thickness, varied depending on the centers. Thus, the CT images obtained from multiple centers were different. Finally, we used the intersection of raw image and the segmentation results as the input to classify COVID-19 pneumonia and other common types of pneumonia in 3D ResNet. Compared to direct classification results when using the original CT images, our segmentation results improved the accuracy of the classification network. This is important in the clinical diagnosis of COVID-19 pneumonia.
Although our TSH-UNet achieved promising segmentation of infected lesions and classification results in COVID-19, there are several limitations in this study. First, segmentation errors may occur in the process of segmentation. There are two main reasons for the segmentation error: one is the TSH-UNet has been verified in CT scans from 6 centers for segmentation and 3 centers for classification. However, the size of dataset is relatively small, resulting in insufficiency of data diversity. In the future, more CT scans from different centers could be added as training and validation dataset to improve the performance of the proposed model. Another one is tiny lesions are inherently not easy to segment, and it is difficult for convolution operations in deep learning to extract the tiny information. We could additionally employ attention mechanism like self-attention to focus specifically on tiny lesions in the future. Besides, early stopping strategy can be considered to prevent network overfitting. Second, for our proposed model, we apply the TSH-UNet to segment the infected regions without classification of types of lung lesions, i.e., GGO, consolidation, or other patterns. In future work, multi-class infection labeling network will be integrated into the current model. Finally, with regard to clinical practice, more detailed clinical information, such as follow-up treatment, was unavailable for validation of the proposed model. Nonetheless, the integration of COVID-19 lesions segmentation and classification in this study allowed for a diagnostic evaluation of the model.
Conclusion
In this study, we proposed a DL-based method TSH-UNet to automatically segment infected regions, which efficiently solved the problem that the 2D network ignores the information of volumetric context and 3D network suffers from high GPU memory consumption and calculation cost. With single-model basis, our method excelled others by a large margin on lung and lesion segmentation. Experiments on multiple centers datasets proved the superiority and the generalization capability of our network. In addition, the results of the segmentation network were used to extract the lesions from the original CT images to classify COVID-19 pneumonia and other common types of pneumonia in an existing classification model (3D ResNet). The classification model achieved the best accuracy with the segmentation results of our TSH-UNet compared with other segmentation network and those obtained through direct classification with the original image, indicating the contribution of our TSH-UNet to the subsequent diagnosis of COVID-19 pneumonia.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge the instrumental and technical support of Multimodal Biomedical Imaging Experimental Platform, Institute of Automation, Chinese Academy of Sciences.
Biographies
Yaxin Shang
is currently a Ph.D. student at School of Computer and Information Technology, Beijing Jiaotong University. Her research interests include computer vision and medical image processing.
Zechen Wei
is currently a Ph.D. student at the Institute of Automation, Chinese Academy of Sciences, China. His research interests include computer vision and medical image processing.
Hui Hui
is a professor at the Institute of Automation, Chinese Academy of Sciences, China. His main research interests include multimodal molecular imaging and biomedical image processing.
Xiaohu Li
is a deputy chief physician of the First Affiliated Hospital of Anhui Medical University. His research interests include cardiovascular imaging and clinical application of new imaging techniques.
Liang Li
works in the Department of Radiology, Renmin Hospital of Wuhan University.
Yongqiang Yu
is a chief physician at the First Affiliated Hospital of Anhui Medical University and vice president of Anhui Medical University. His research interests include neuroimaging, molecular imaging and new imaging technologies.
Ligong Lu
is a chief physician at Zhuhai Interventional Medical Center, Zhuhai Precision Medical Center and Zhuhai Hospital Affiliated with Jinan University.
Li Li
is a radiologist in the Department of Radiology, Beijing Youan Hospital, Capital Medical University, Beijing, China.
Hongjun Li
is a chief physician at Department of Radiology, Beijing Youan Hospital, Capital Medical University, Beijing, China.
Qi Yang
is a chief physician at Department of Radiology, Beijing Chaoyang Hospital, Capital Medical University, Beijing, China.
Meiyun Wang
is a chief physician at Department of Medical Imaging, Henan Provincial People’s Hospital & the People’s Hospital of Zhengzhou University.
Meixiao Zhan
is an associate professor at Zhuhai Interventional Medical Center, Zhuhai Precision Medical Center, Zhuhai Hospital Affiliated with Jinan University, Zhuhai, Guangdong, China.
Wei Wang
is currently a M.D. student at Chinese PLA Medical School, China.
Guanghao Zhang
is currently a M.S. student at the Institute of Automation, Chinese Academy of Sciences, China.
Xiangjun Wu
is currently a Ph.D. student at Beihang University, China. His research interests include computer vision and medical image processing.
Li Wang
is a physician at the Department of Infection Prevention and Control Office, Renmin Hospital of Wuhan University, Wuhan, China.
Jie Liu
is a professor at School of Computer and Information Technology, Beijing Jiaotong University, Beijing, China. His research interests are medical image processing, artificial intelligence and biomedical applications.
Jie Tian
is a professor at the Beijing Advanced Innovation Center for Big Data-Based Precision Medicine, School of Medicine, Beihang University, China. His research interests include medical image process and molecular imaging.
Yunfei Zha
is a chief physician and director of the Department of Radiology, Renmin Hospital of Wuhan University.
Funding
This work was supported in part by the National Key Research and Development Program of China (2017YFA0700401, 2019YFC0118100); Novel Coronavirus Pneumonia Emergency Key Project of Science and Technology of Hubei Province (No.2020FCA015); National Natural Science Foundation of China (81671851, 81827808, 81571836, 61936013, Z211100003521003); CAS Youth Innovation Promotion Association under Grant (2018167) and CAS Key Technology Talent Program. The Project of High-Level Talents Team Introduction in Zhuhai City (Zhuhai HLHPTP201703).
Declarations
Ethics approval
All human and animal studies have been approved by the appropriate ethics committee and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yaxin Shang, Zechen Wei, Hui Hui, Xiaohu Li, and Liang Li contributed equally to this work.
Contributor Information
Yaxin Shang, Email: 20112039@bjtu.edu.cn.
Zechen Wei, Email: weizechen2019@ia.ac.cn.
Hui Hui, Email: hui.hui@ia.ac.cn.
Xiaohu Li, Email: 13955168568@126.com.
Liang Li, Email: 15001017169@139.com.
Yongqiang Yu, Email: cjr.yuyongqiang@vip.163.com.
Ligong Lu, Email: lu_ligong@163.com.
Li Li, Email: liliang_082@163.com.
Hongjun Li, Email: lihongjun00113@126.com.
Qi Yang, Email: yangyangqiqi@gmail.com.
Meiyun Wang, Email: mywang@ha.edu.cn.
Meixiao Zhan, Email: zhanmeixiao1987@126.com.
Wei Wang, Email: avigor@163.com.
Guanghao Zhang, Email: zhangguanghao19@mails.ucas.ac.cn.
Xiangjun Wu, Email: wuxiangjun18@mails.ucas.ac.cn.
Li Wang, Email: toto-86@163.com.
Jie Liu, Email: jieliu@bjtu.edu.cn.
Jie Tian, Email: tian@ieee.org.
Yunfei Zha, Email: zhayunfei999@126.com.
References
- 1.Li JY, et al. The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future. Microbes Infect. 2020;22(2):80–85. doi: 10.1016/j.micinf.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benvenuto D, et al. The global spread of 2019-nCoV: a molecular evolutionary analysis. Pathog Glob Health. 2020;114(2):64–67. doi: 10.1080/20477724.2020.1725339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020) Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology 296(2):E41–E45 [DOI] [PMC free article] [PubMed]
- 4.Chung M, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D et al (2020) Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA323(11):1061–1069 [DOI] [PMC free article] [PubMed]
- 6.Li C et al (2020) Classification of severe and critical Covid-19 using deep learning and radiomics. IEEE J Biomed Health Inform 24(12):3585–3594 [DOI] [PMC free article] [PubMed]
- 7.Singh M, Bansal S, Ahuja S, Dubey RK, Panigrahi BK, Dey N. Transfer learning–based ensemble support vector machine model for automated COVID-19 detection using lung computerized tomography scan data. Med Biol Eng Compu. 2021;59(4):825–839. doi: 10.1007/s11517-020-02299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamshidi M, et al. Artificial intelligence and COVID-19: deep learning approaches for diagnosis and treatment. IEEE Access. 2020;8:109581–109595. doi: 10.1109/ACCESS.2020.3001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng L, et al. A deep learning prognosis model help alert for COVID-19 patients at high-risk of death: a multi-center study. IEEE J Biomed Health Inform. 2020;24(12):3576–3584. doi: 10.1109/JBHI.2020.3034296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X et al (2020) A deep learning system to screen novel coronavirus disease 2019 pneumonia. Eng 6(10):1122–1129 [DOI] [PMC free article] [PubMed]
- 11.Wu X et al (2020) Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur J Radiology 128:109041 [DOI] [PMC free article] [PubMed]
- 12.Alshalan R, Al-Khalifa H, Alsaeed D, Al-Baity H, Alshalan S (2020) Detection of hate speech in COVID-19-related tweets in the Arab Region: deep learning and topic modeling approach. J Med Internet Res 22(12):e22609 [DOI] [PMC free article] [PubMed]
- 13.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang MJ et al (2020) CT radiomics can help screen the Coronavirus disease 2019 (COVID-19): a preliminary study. Sci China Inform Sci63:172103
- 15.Dong D et al (2021) The role of imaging in the detection and management of COVID-19: a review. IEEE Rev Biomed Eng 14:16–29 [DOI] [PubMed]
- 16.Khan S, Huh J, Ye JC (2020) Adaptive and compressive beamforming using deep learning for medical ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control 67(8):1558–1572 [DOI] [PubMed]
- 17.Shan F et al (2021) Abnormal lung quantification in chest CT images of COVID-19 patients with deep learning and its application to severity prediction. Med Phys 48(4):1633–1645 [DOI] [PMC free article] [PubMed]
- 18.Wang S et al (2021) A deep learning algorithm using CT images to screen for corona virus disease (COVID-19). Eur Radiol 31(8):6096–6104 [DOI] [PMC free article] [PubMed]
- 19.Huang L, et al. Serial quantitative chest CT assessment of COVID-19: a deep learning approach. Radiol Cardiothorac Imaging. 2020;2(2):e200075. doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amyar A, Modzelewski R, Li H, Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: classification and segmentation. Comput Biol Med. 2020;126:104037. doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou L, et al. A rapid, accurate and machine-agnostic segmentation and quantification method for CT-based COVID-19 Diagnosis. IEEE Trans Med Imaging. 2020;39(8):2638–2652. doi: 10.1109/TMI.2020.3001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gianchandani N, Jaiswal A, Singh D, Kumar V, Kaur M (2020) Rapid COVID-19 diagnosis using ensemble deep transfer learning models from chest radiographic images. J Amb Intel Hum Comp 1–13 [DOI] [PMC free article] [PubMed]
- 23.Li L et al (2020) Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology 296(2):E65–E71 [DOI] [PMC free article] [PubMed]
- 24.Fan DP et al (2020) Inf-Net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans Med Imaging 39(8):2626–2637 [DOI] [PubMed]
- 25.Gao K et al (2020) Dual-branch combination network (DCN): towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med Image Anal 67:101836 [DOI] [PMC free article] [PubMed]
- 26.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. Proc IEEE conf Comput Vis Pattern Recognit, 2016, pp. 770–778
- 27.Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imaging. 2018;37(12):2663–2674. doi: 10.1109/TMI.2018.2845918. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: a nested U-Net architecture for medical image segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support. 2018;2018(11045):3–11. doi: 10.1007/978-3-030-00889-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hara K, Kataoka H, Satoh Y (2018) Can spatiotemporal 3d cnns retrace the history of 2d cnns and imagenet?. Proc IEEE conf Comput Vis Pattern Recognit 2018:6546–6555
- 30.Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans Med Imaging. 2020;39(6):1856–1867. doi: 10.1109/TMI.2019.2959609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kline DM, Berardi VL. Revisiting squared-error and cross-entropy functions for training neural network classifiers. Neural Comput Appl. 2005;14(4):310–318. doi: 10.1007/s00521-005-0467-y. [DOI] [Google Scholar]
- 32.Cai C, et al. Transfer learning for drug discovery. J Med Chem. 2020;63(16):8683–8694. doi: 10.1021/acs.jmedchem.9b02147. [DOI] [PubMed] [Google Scholar]
- 33.Taylor ME, Stone P (2009) Transfer learning for reinforcement learning domains: a survey. J Mach Learng Res 10(7):1633–1685
- 34.Khan S, Islam N, Jan Z, Din IU, Rodrigues JJC. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recogn Lett. 2019;125:1–6. doi: 10.1016/j.patrec.2019.03.022. [DOI] [Google Scholar]
- 35.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. Proc IEEE conf Comput Vis Pattern Recognit. 2261–2269
- 36.Jha S, Kumar R, Priyadarshini I, Smarandache F, Long HV. Neutrosophic image segmentation with dice coefficients. Measurement. 2019;134:762–772. doi: 10.1016/j.measurement.2018.11.006. [DOI] [Google Scholar]
- 37.Beauchemin M, Thomson KP, Edwards G. On the Hausdorff distance used for the evaluation of segmentation results. Can J Remote Sens. 1998;24(1):3–8. doi: 10.1080/07038992.1998.10874685. [DOI] [Google Scholar]
- 38.Pham D, Xu C, Prince J (2000) Current methods in medical image segmentation. Annu Rev Biomed Eng 2(1):315–337 [DOI] [PubMed]
- 39.Ma J et al (2020) Toward data-efficient learning: A benchmark for COVID-19 CT lung and infection segmentation. Med Phys 48(3):1197–1210 [DOI] [PubMed]
- 40.Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(4):640–651. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 41.Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(12):2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 42.Wang B, Jin S, Yan Q, Xu H, Dong J (2020) AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system. Appl Soft Comput 98:106897 [DOI] [PMC free article] [PubMed]
- 43.Müller D, Soto-Rey I, Kramer F (2020) Robust chest CT image segmentation of COVID-19 lung infection based on limited data. Inform Med Unlocked 25:100681 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.