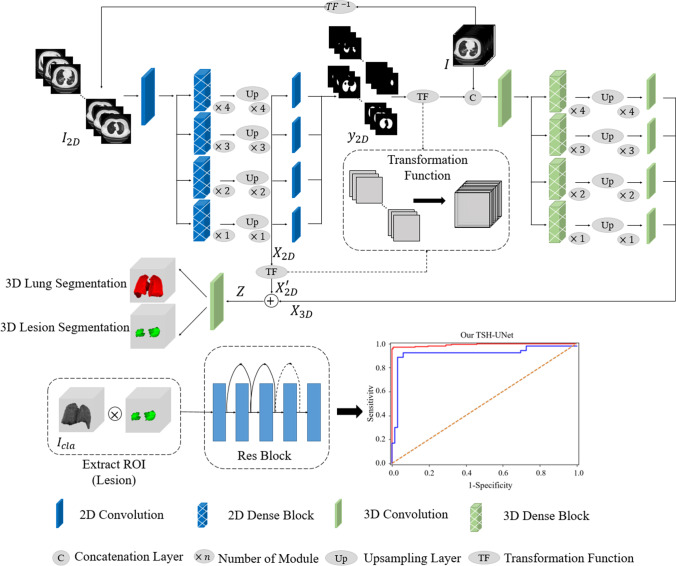
Fig. 1.
Main framework of the proposed model. The structure of TSH-UNet for lung and lesion segmentation includes a 2D network and a 3D network, the detailed architecture of which can be found in supplementary Fig. 1 and supplementary Table 1. The input volume data are transformed into three consecutive slices and fed into the 2D network, yielding a coarse segmentation result. Then, after concatenation with the predicted volumes from 2D network, the input volume data are fed into 3D network to extract interslice features. Finally, the hybrid features are jointly optimized in the 3D network to accurately segment lungs and lesions. For classification, the region of interest is extracted using the segmentation result. The 3D network used in the classification part of our model is adapted from 3D ResNet. Finally, the model outputs the probability of COVID-19 pneumonia and common pneumonia. The 2D network consisted of 167 layers, including convolutional layers, pooling layers, upsampling layers, transition layers, and a dense block. The dense block represented the cascade of several microblocks, where all layers were directly connected. The transition layers were used to resize the feature maps, which were composed of a batch normalization layer and convolutional layer (1 × 1) followed by an average pooling layer. We set the compression factor as 0.5 in the transition layers to prevent the feature maps from expanding. Bilinear interpolation was employed in the upsampling layer, followed by the sum of low-level features and a convolutional layer (3 × 3). In addition, batch normalization and the rectified linear unit were used before each convolutional layer in the architecture