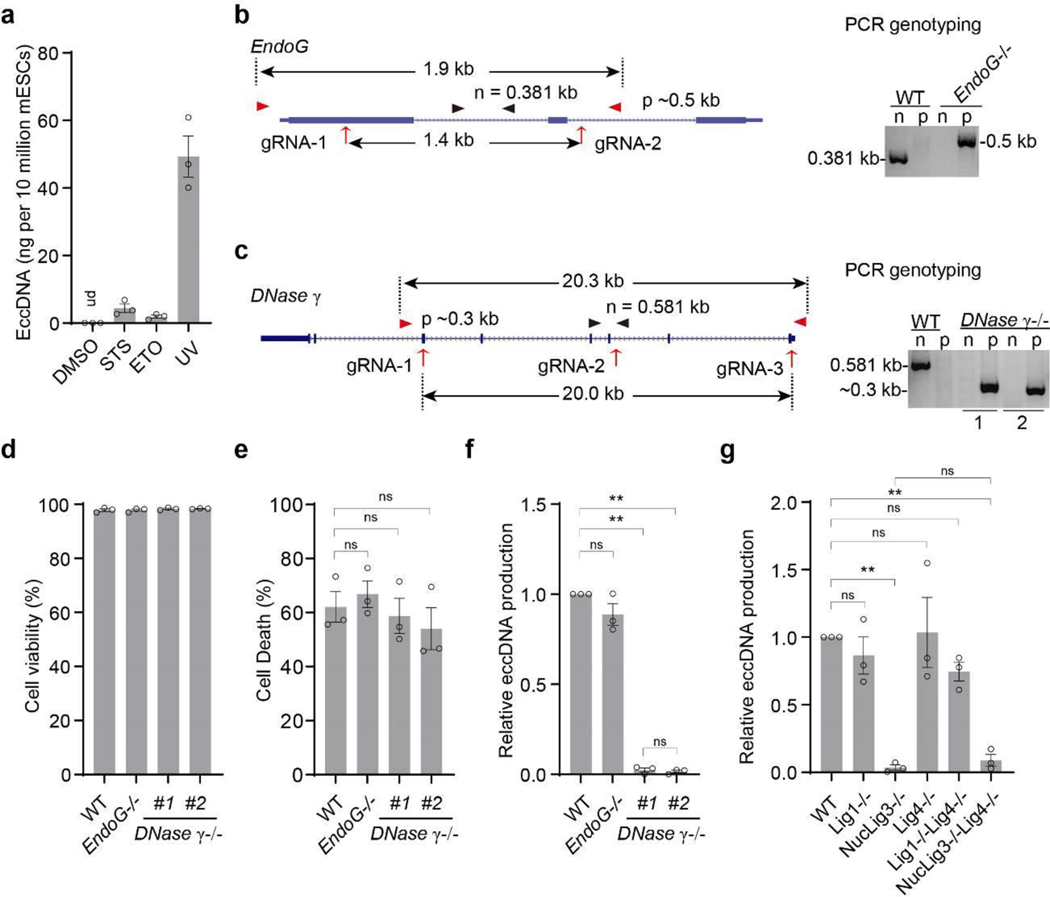
Extended Data Fig. 4. Effects of Endonuclease G, DNase γ, and DNA ligases knockout on apoptosis and eccDNA generation.
a, Quantification of eccDNA from cells of the indicated treatment. EccDNA production were presented as nanogram per 10 million cells; ud, under detection limit. Bars indicate mean ± SEM of three independent experiments.
b, Diagram illustration and PCR confirmation of the EndoG knockout mESC line. Gene structure of EndoG, sgRNAs (red arrows), two sets of screen primers for internal sites (black arrowhead, “n” =negative knockout cell line) and external sites (red arrowhead, “p” =positive knockout cell line) were shown.
c, Diagram illustration and PCR confirmation of the DNase γ knockout mESC lines. Gene structure of DNase γ, sgRNAs (red arrows), two sets of screen primers for internal sites (black arrowhead, “n” =negative knockout cell line) and external sites (red arrowhead, “p” =positive knockout cell line) were shown.
d, Cell viability is not affected by EndoG or DNase γ KO. Cell viability was evaluated by flow cytometry, after staining with Far Red Live/Dead Cell Stain Kit, FSC-A/SSC-A and FSC-A/FSC-H were sequentially used to exclude debris and gate on singlets, respectively, then gate on APC+ as dead cells. Bars indicate mean ± SEM. of dead cell ratio from three independent experiments.
e, Deficiency of DNase 𝛾 or EndoG in mESC do not significantly alter UV-induced cell death. Cell death was measured by flow cytometry, after staining with Far Red Live/Dead Cell Stain Kit, FSC-A/SSC-A and FSC-A/FSC-H were sequentially used to exclude debris and gate on singlets, respectively, then gate on APC+ as dead cells. Bar indicate mean ± SEM. of dead cell ratio of 3 independent experiments.
f, Quantification of eccDNAs presented in Fig. 3d. EccDNAs below the mtDNA band were quantified by densitometry (by Image J 1.53e) and presented as relative levels to that in WT cells of parallel eccDNA purifications. Bars indicate mean ± SEM. of three independent experiments.
g, Quantification of eccDNA below the mtDNA band by densitometry (by Image J 1.53e). Data is presented as relative levels to that purified from WT cells of parallel eccDNA purifications (Fig. 3g), bars indicate mean ± SEM of three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparison test was used for multiple comparisons in e, f, g. **, p<0.01; ns, no significant.