Figure 1.
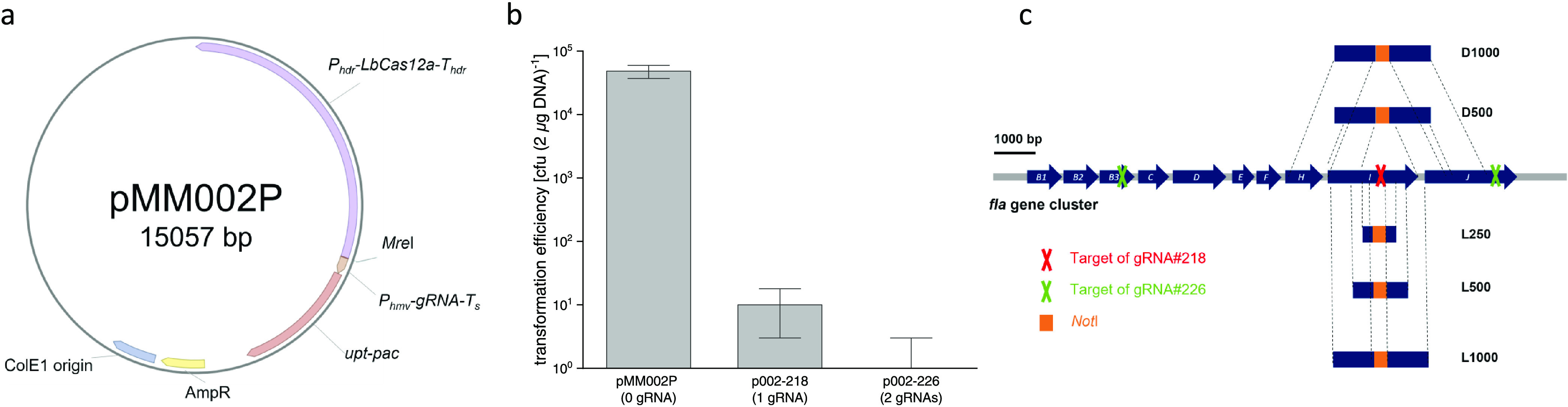
General features of the CRISPR/LbCas12a genome editing. (a) Genetic map of the CRISPR/LbCas12a pMM002P plasmid. The M. maripaludis S2 uracil phosphoribosyltransferase gene (upt), which serves as a counter-selective marker, and the codon-optimized puromycin N-acetyltransferase gene (pac) are coexpressed via the Pmcr promoter.4 LbCas12a expression is driven by the Phdr promoter from Methanococcus voltae A3. gRNA expression is driven by the M. voltae A3 Phmv histone promoter. Two PaqCI sites between the direct repeat sequence and the synthetic terminator in the opposite direction for spacer fusion is used for gRNA insertion (not displayed). The gRNA of the plasmid pMM002P that contains two PaqCI sites does not target the chromosome. An MreI restriction site assigned between the gRNA and Cas elements is used for RF insertion. (b) CRISPR/LbCas12a triggered DSBs. Shown are the transformation efficiencies [cfu (2 μg DNA)−1] for the CRISPR/LbCas12a pMM002P plasmids with one, two, or no gRNAs that were used to transform M. maripaludis. Error bars represent the standard deviation of the values obtained for the transformation efficiency (n = 3). (c) Schematic outline of the repair fragment edits. A NotI site is placed between the two homologous arms.
