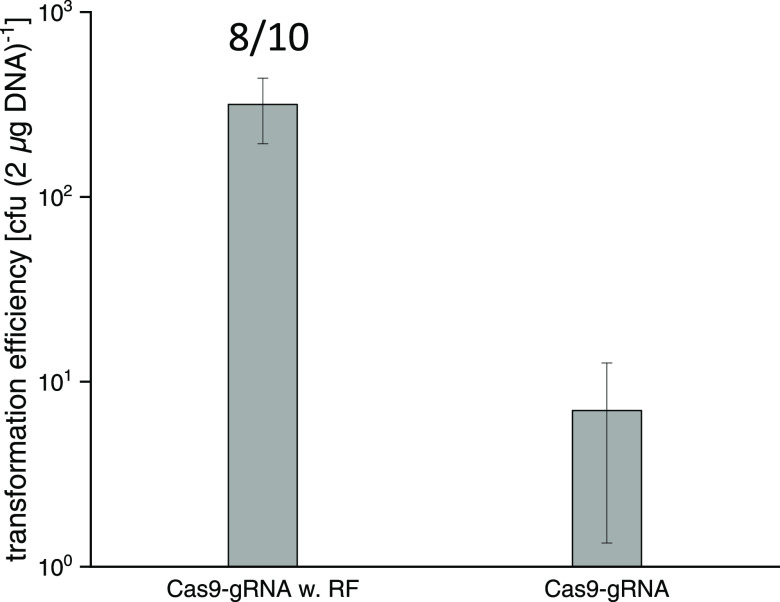
Figure 4.
CRISPR/SpCas9 genome-edited replacement of the M. maripaludis alanine dehydrogenase–alanine racemase (ald-alr 1.9 kbp) genes with a 4.2 kbp fragment. Shown are the transformation efficiencies [cfu (2 μg DNA)−1] for the CRISPR/SpCas9 plasmids that were used to transform M. maripaludis. The left bar indicates that the cells were transformed with a suicide plasmid containing the integration cassette and a CRISPR/SpCas9 plasmid carrying a gRNA targeting to the ald-alr. The right bar indicates that the cells were only transformed with a CRISPR/SpCas9 plasmid carrying a gRNA targeting to the ald-alr. Error bars represent the standard deviation of the values obtained for the transformation efficiency (n = 3). The positive rate representing the fraction of correctly edited colonies per colonies tested by PCR is shown for the transformations (numbers above the left bar).