Figure 1.
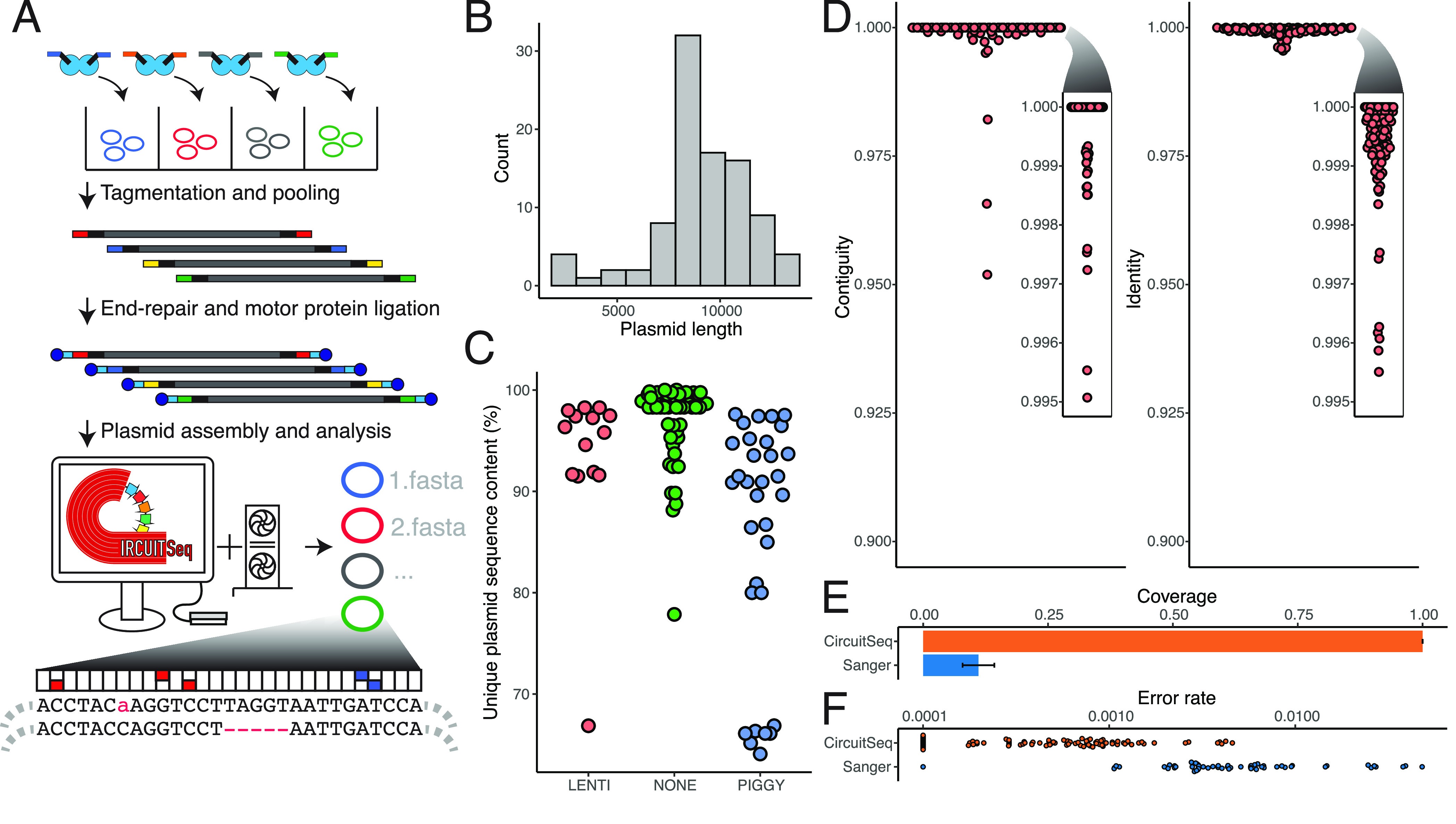
(A) Schematic representation of Circuit-seq. Plasmids are arranged in a 96-well plate and are tagmented with well-specific barcodes. Samples are then pooled, end-repaired, adapter-ligated, and processed on an Oxford Nanopore Flongle flow cell. The generated data are then run through a custom NextFlow pipeline, producing the final assembled sequences. (B) distribution of plasmid sizes used in our experiment. (C) Unique (nonrepetitive) DNA fraction for our input plasmid pool. Lentiviral (LENTI) and PiggyBac (PIGGY) payload plasmids have more repetitive sequence in their backbones, a key criterion to challenge our assembly pipeline. (D) Contiguity and identity scores for the polished assemblies were calculated by comparison to the known reference. (E) Proportions of the full plasmid covered by Circuit-seq assemblies and Sanger sequencing. (F) Error rates for Circuit-seq assemblies and Sanger sequencing calculated by comparison with the known reference. Sanger sequences were prefiltered to remove sequences with >10% error to exclude technical errors from the analysis.
