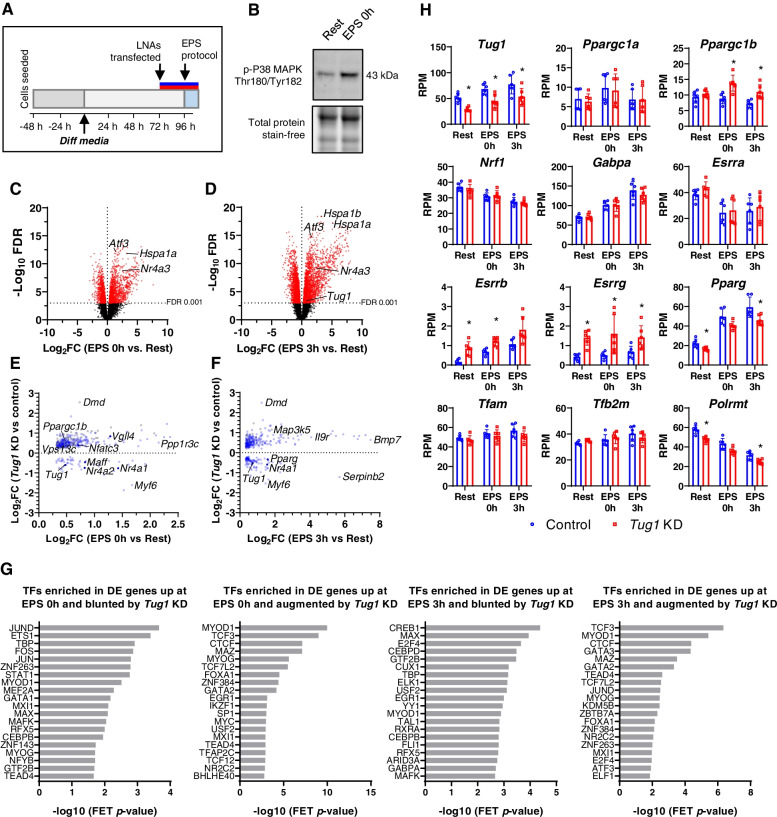
Fig. 7.
Tug1 KD modifies transcriptomic responses to simulated exercise in myotubes. A C2C12 myocytes were differentiated for 3 days, transfected with Tug1 or control LNAs for 24 h, then on day 4 underwent electrical pulse stimulation (EPS) or rest for 3 h before being harvested immediately (EPS 0h) or after 3-h recovery (EPS 3h) for RNA-seq. B Representative immunoblot of phosphorylated p38 MAPK immediately after 3 h EPS. C RNA-seq volcano plot of genes in control myotubes differentially expressed immediately after the EPS protocol, and D 3 h after EPS compared to Rest (red points, FDR<0.001). E Scatterplot of genes that were upregulated (FDR<0.05) at EPS 0h or F EPS 3h compared to Rest, but blunted or augmented by Tug1 KD compared to control (FDR<0.05); grey points represent genes upregulated only in Tug1 KD myotubes after EPS but that were not upregulated in control myotubes after EPS. H RNA-seq data in reads per million (RPM) for Tug1 and selected genes associated with mitochondrial biogenesis. G Transcription factors enriched in genes (shown in panels E and F) that were upregulated by EPS yet blunted or augmented due to Tug1 KD. FET, Fisher’s exact test. Data are from n=6 libraries per group (triplicate wells from two independent experiments for each condition); *FDR<0.05 Tug1 KD vs. NC. See also Additional file 1: Supplementary Fig. S5, as well as Additional file 2: Supplementary Tables S3 to S9