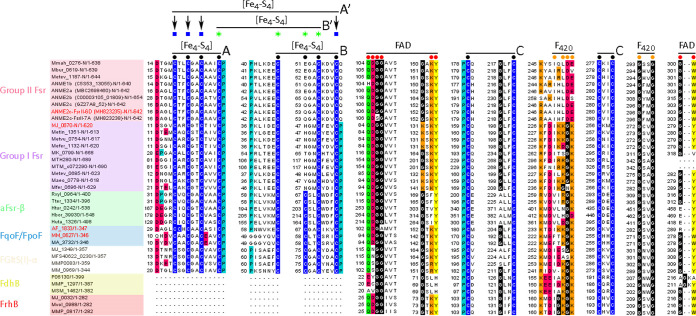
FIG 4.
Multiple primary sequence alignment for ANME2c-FsrII-6D-N homologs. Conserved amino acid residues are represented as follows: blue, cysteine; cyan, proline; black, glycine; red, negatively charged (D or E); green, polar uncharged (S, T, N, or Q); orange, positively charged (H, R, or K); yellow, aromatic (F, Y, or W). Black bullets with overlines and A and B notations show [Fe4-S4] cluster assembly motifs; black circles with overlines and C notation represent conserved cysteine residues, red circles with overlines show the flavin binding motif (89, 90), and orange circles with overlines show the F420 binding motif (89); blue squares and green stars with overlines and A′ and B′ notations, respectively, show [Fe4-S4] cluster assembly motifs identified in the AlphaFold2 models. Fsr, F420-dependent sulfite reductases (14, 19, 22); aFsr-β, F420H2 dehydrogenase subunit of a putative F420H2-dependent assimilatory type siroheme sulfite reductase (22); FqoF/FpoF, F420H2 dehydrogenase subunit of a membrane-bound proton pumping F420H2 dehydrogenase complex (32, 33); FGltS(I)-α, F420H2 dehydrogenase subunit of a putative F420H2-dependent glutamate synthase (22, 91); FdhB, formate dehydrogenase subunit B (92); FrhB: F420-reducing [NiFe]-dehydrogenase subunit B (77). Open reading frame (ORF) numbers of the proteins, except those from ANME, are presented in the NCBI format (abbreviated organism name_respective ORF number). For an ANME protein, the NCBI ORF number appears within parentheses following the abbreviated organism name. “-N” indicates that only the N-terminal part of the polypeptide is shown. Ranges following slashes are amino acid residues for the complete protein. The names of proteins that are of particular interest in the study are shown in red. ANME2c-FsrII-6D-N, MH823235; MjFsrI-N, MJ_0870; FpoF of Archaeoglobus fulgidus, AF_1833; FqoF of Methanosarcina mazei, MM_0627. Abbreviations of organism names: Mmah, Methanohalophilus mahii DSM 5219; Mbur, Methanococcoides burtonii DSM 6242; Metev, Methanohalobium evestigatum Z-7303; MJ, Methanocaldococcus jannaschii DSM 2661; Metin, Methanocaldococcus infernus ME; Metvu, Methanocaldococcus vulcanius M7; Mefer, Methanocaldococcus fervens AG86; MK, Methanopyrus kandleri AV19; MTH, Methanothermobacter thermautotrophicus ΔH; MTM, Methanothermobacter marburgensis strain Marburg; Maeo, Methanococcus aeolicus Nankai-3; Mfer, Methanothermus fervidus DSM 2088; Rxyl, Rubrobacter xylanophilus DSM 9941; Tter, Thermobaculum terrenum ATCC BAA-798; Htur, Haloterrigena turkmenica DSM 5511; Hbor, Halogeometricum borinquense DSM 11551; Huta, Halorhabdus utahensis DSM 12940; AF, Archaeoglobus fulgidus DSM 4304; MM, Methanosarcina mazei Gö1; MA, Methanosarcina acetivorans C2A; MFS40622, Methanocaldococcus sp. FS406-22; MMP, Methanococcus maripaludis S2; P06130, accession number for Methanobacterium formicicum FdhB; MSM, Methanobrevibacter smithii ATCC 35061; Mvol, Methanococcus voltae A3; ANME, anaerobic methanotrophic archaea.