FIG 6.
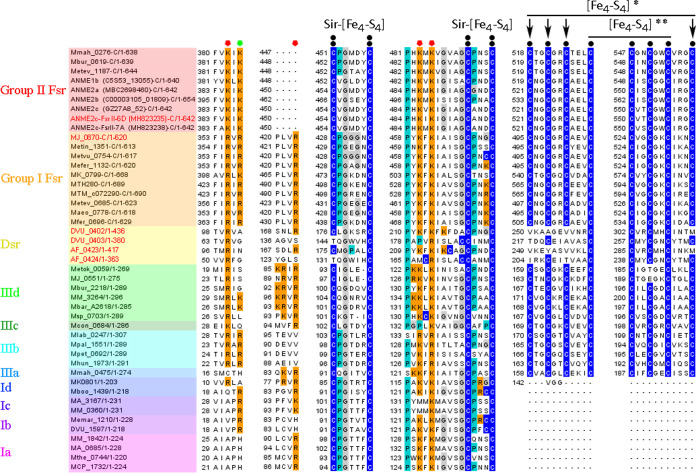
Multiple-sequence alignment of ANME2c-FsrII-6D-C homologs. Amino acid residues are represented as follows: blue, cysteine; cyan, proline; gray, glycine; orange, positively charged (H, R, or K). Black circles represent conserved cysteine residues, double black bullets represent siroheme-[Fe4-S4] cluster binding residues, red polygons show sulfite binding residues, the green polygon shows conserved positively charged residues [RK] in group I and group II Fsrs, black bullets with overlines show [Fe4-S4] cluster assembly motifs, and single and double asterisks show peripheral and additional [Fe4-S4] centers, respectively (22). ORF numbers of the proteins are presented as described in the legend to Fig. 4. “-C” indicates that only the C-terminal part of the polypeptide is shown. Range following slashes are amino acid residues for the complete protein. Dsr, dissimilatory sulfite reductase; Ia-d, group I dissimilatory sulfite reductase like protein (Dsr-LP) subgroups a to d; IIIa-d, group III Dsr-LP subgroups a to d. The names of proteins that are of particular interest in the study are shown in red. ANME2c-FsrII-6D-C, MH823235; MjFsrI-C, MJ_0870; Dv-DsrA/B, Dsr subunits A and B of Desulfovibrio vulgaris strain Hildenborough, DVU_0402 and DVU_0403; Af-DsrA/B, Dsr subunits A and B of Archaeoglobus fulgidus DSM 4304, AF_0423 and AF_0424. Metok, Methanothermococcus okinawensis IH1; Mbar, Methanosarcina barkeri strain Fusaro; Msp, Methanosphaera stadtmanae DSM 3091; Mcon, Methanosaeta concilii GP-6; Mlab, Methanocorpusculum labreanum Z; Mpal, Methanosphaerula palustris E1-9c; Mpet, Methanoplanus petrolearius DSM 11571; Mhun, Methanospirillum hungatei JF-1; Mboo, Candidatus Methanoregula boonei 6A8; Memar, Methanoculleus marisnigri JR1; Mthe, Methanosaeta thermophila PT; MCP, Methanocella paludicola SANAE. The legend to Fig. 4 presents abbreviations of other organisms’ names.