FIGURE 2.
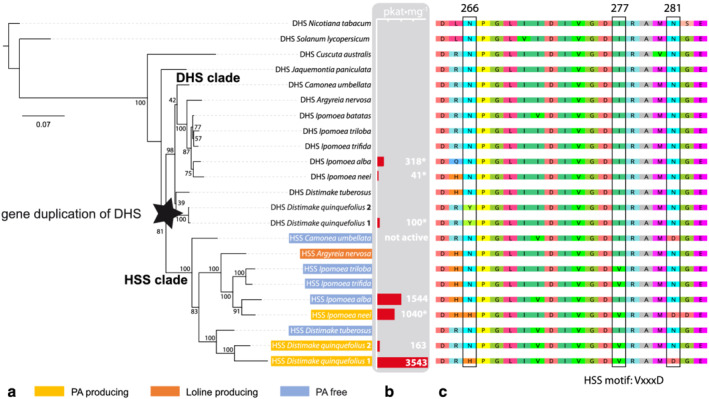
Phylogenetic analyses of deoxyhypusine synthase (DHS)‐ and homospermidine synthase (HSS)‐encoding sequences. (a) A maximum likelihood tree was built from the cDNA sequences of HSS and DHS from the Convolvulaceae family and two DHS sequences from Solanaceae family. Percentages of bootstrap support values (for 1000 replicates) are indicated. The ancient duplication is highlighted, and the DHS and HSS clades are indicated. (b) Selected sequences were chosen for biochemical analyses of the encoded enzymes. The calculated specific activity (pkat·mg) of homospermidine production of these enzymes in glycine‐based assay buffer is given (see Table 3). Specific activities marked with an asterisk were taken from a previous study (Kaltenegger et al., 2013) (c) Alignment of the functionally characterized “HSS motif” (H‐V‐D)
