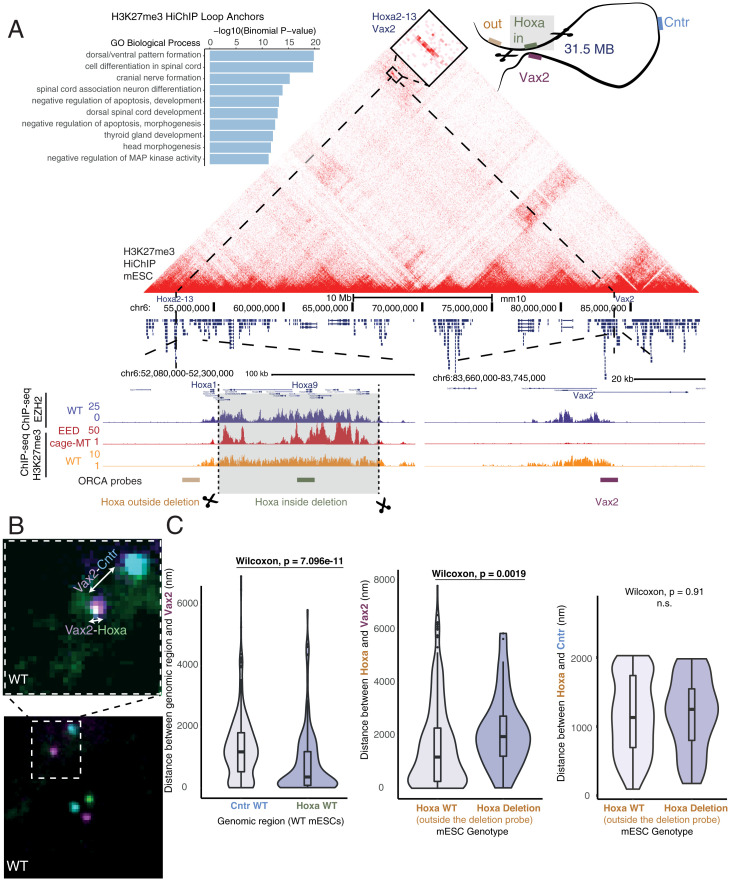
Fig. 2.
Deletion of the H3K37me3-associated loop anchor at the Hoxa cluster alters long-range 3D interactions. (A) Gene Ontology terms enriched at mESC H3K27me3-associated loop anchors (Left). H3K27me3 HiChIP contact matrix (10-kb resolution) visualizing Polycomb-associated interactions at the 40-Mb region encompassing the Hoxa cluster and Vax2 in mESCs. ChIP-seq signal for WT EZH2 and H3K27me3 (WT and EED cage MT) and the position of ORCA probes are shown below. (B) ORCA imaging of three probes targeting Hoxa, Vax2, and control (Cntr) regions demonstrates the interaction between the Hoxa cluster with Vax2 at the single-nucleus level in WT mESCs. Violin plots of the distance between Vax2 and the Hoxa or Cntr probes (WT cells, n = 2,190; MT cells, n = 520). The Hoxa probe located within the Polycomb-associated loop anchor was used in WT mESCs. (C) Violin plots of the distance between Hoxa and Vax2 (as measured by ORCA) for WT and Hoxa deletion mESCs (Left). Violin plots of the distance between Hoxa and Cntr (as measured by ORCA) for WT and Hoxa deletion mESCs (Right). As the Hoxa Polycomb-associated loop anchor is deleted in Hoxa deletion mESCs, a probe targeting a region adjacent to the Hoxa loop anchor was used for both WT and Hoxa deletion mESCs. Wilcoxon test was used for significance. n.s., not significant.