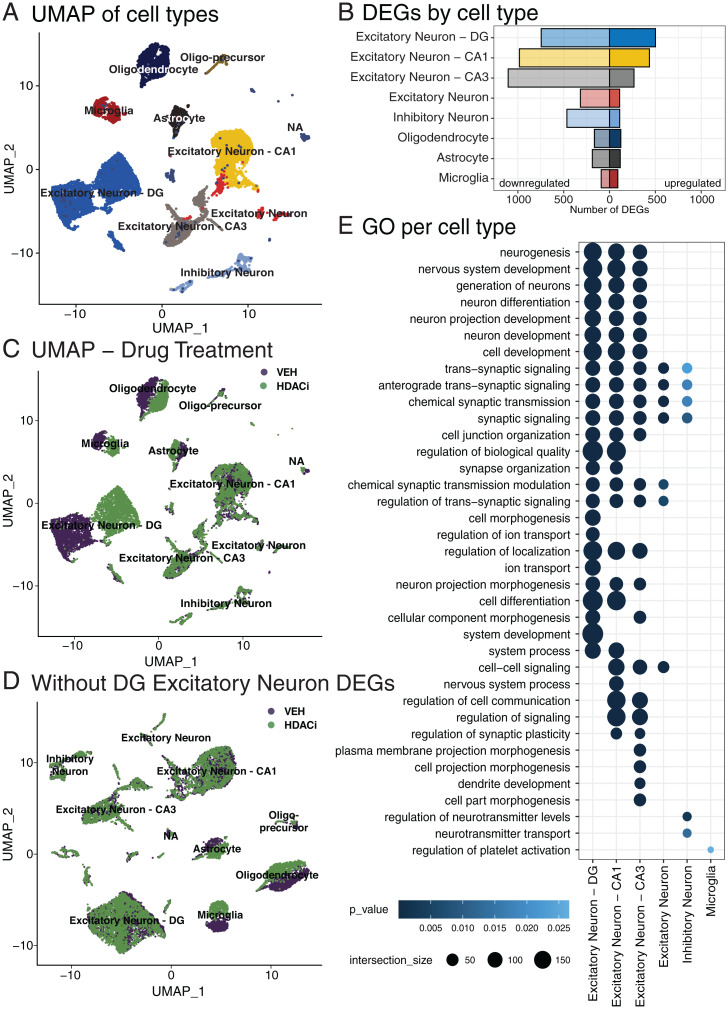
Fig. 3.
HDACi activates different transcriptional cascades within hippocampal cell types. (A) UMAP of 15,339 nuclei from the full hippocampus colored by 10 identified cell types. (B) Number of up-regulated (Right; log2FC ≥ 1; false-discovery rate [FDR] ≤ 0.05) and down-regulated (Left; log2FC ≤ −1; FDR ≤ 0.05) genes in each cell type that has significant GOs. (C) UMAP of nuclei colored by sample drug treatment. (D) UMAP, colored by drug treatment, after removing the 501 up-regulated genes in the DG excitatory neurons and reclustering. (E) Top 25 enriched GOs of genes up-regulated after HDACi treatment and CFC in each cell type. Cell types that have no ontologies are not represented. n = 2 biological replicates per group (HDACi-CFC and VEH-CFC).