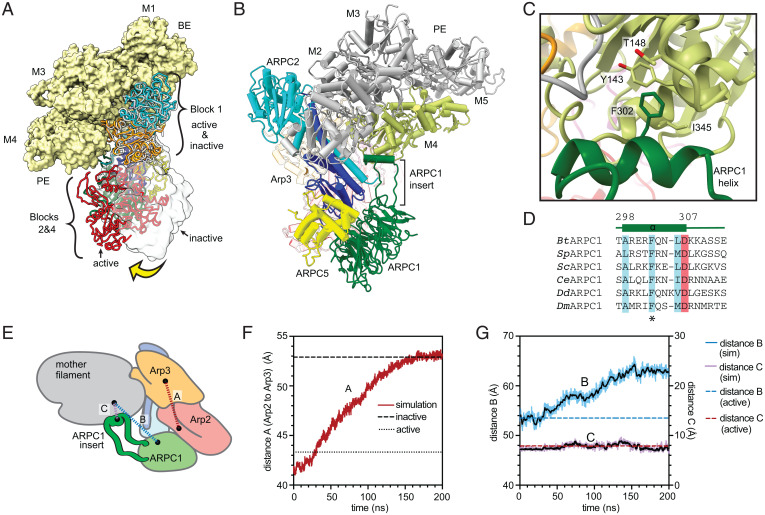
Fig. 6.
Interaction of Arp2/3 complex with actin filaments may not stimulate movement of Arp2 into the short pitch conformation. (A) Comparison of mother filament binding contacts of the activated (flattened, short pitch) Arp2/3 complex to those of the inactive (splayed, twisted) Arp2/3 complex (PDB 4JD2, rendered in gray ribbon or transparent gray surface). The inactive Arp2/3 complex was modeled onto the mother filament by superposing block 1 onto block 1 in the branch junction model. Yellow arrow shows movement of blocks 2 and 4 stimulated by clamp twisting. Block 3 is omitted for clarity. (B) Ribbon diagram of a portion of the branch junction model highlighting the interaction between the ARPC1 protrusion helix and the mother filament. (C) Interaction of the ARPC1 protrusion helix with subunit M4 in the mother filament. (D) Sequence alignment showing conservation of a subset of hydrophobic (cyan) and acidic (red) residues in the ARPC1 protrusion helix. (E) Schematic showing key distances in molecular dynamics (MD) simulations plotted in F and G. (F) Plot of distance A, which measures the distance between the centers of mass of Arp2 and Arp3 (subdomains 3 and 4, see Materials and Methods), versus time during the simulation. The equilibrated branch junction structure is pulled to the splayed conformation over 150 ns via a biased MD simulation. The distance between Arp2 and Arp3 in the active (short pitch, flattened) conformation in the branch junction is shown with a dotted line, while the distance in the inactive (splayed, twisted) conformation is shown with a dashed line. (G) Plot of distances B and C, which measure the distance between the globular portion of ARPC1 (distance B) or the ARPC1 protrusion helix (distance C) and the mother filament versus time during the simulation.