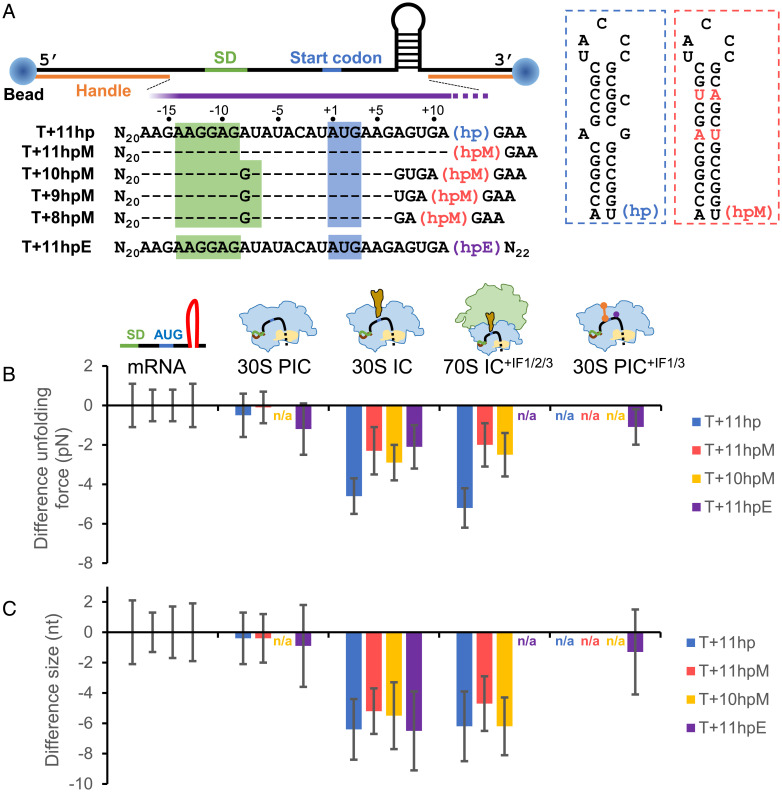
Fig. 5.
The ribosome can unwind up to 3 bp of the downstream mRNA structure after the binding of initiator tRNA. (A) Experimental setup for optical tweezers. An anti-digoxigenin antibody–coated polystyrene bead (Left) and a streptavidin-coated bead (Right) were attached to the ends of mRNA through the digoxigenin- and biotin-labeled DNA handles, respectively. The mRNA sequences (5′ to 3′) between the two handles are shown. Sequences identical to T+11hp are denoted as dashes for clarity. The approximate ribosome footprint is indicated by the purple bar, which overlaps part of the downstream hairpins (hp, hpM, and hpE). In hpM (red boxed), the mismatched and bulged bases of hp (blue boxed) were changed such that the whole stem was correctly base paired. For hpE, see SI Appendix, Fig. S4A for detail. (B and C) Difference in unfolding force (B) and size (C) of the downstream hairpins. For each of the indicated mRNA constructs, the unfolding force and size of the hairpin in the designated ribosome-binding states (30S PIC, 30S IC, 70S IC, and 30S PIC+IF1/3) were plotted as the difference from that of the mRNA alone. Error bars represent SDs; n/a, not available.