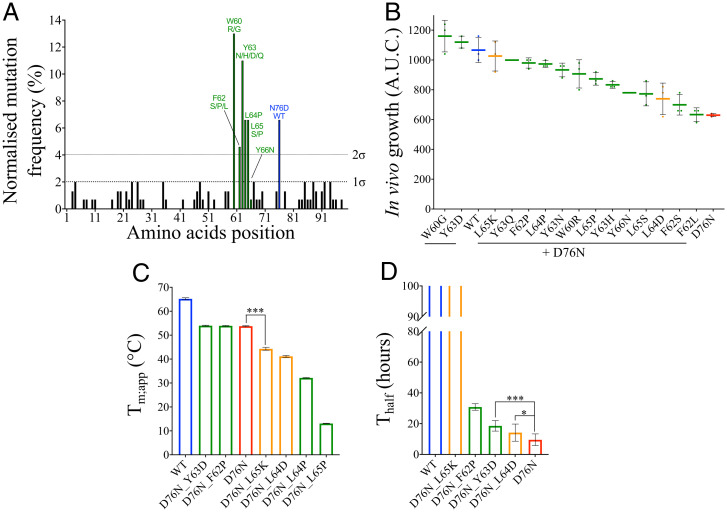
Fig. 5.
Evolving D76N-β2m to improve its properties. (A) Frequency of amino acid substitutions for the 56 unique sequences during screening of the D76N-β2m* library reveals the APR (residues 60 to 66, in green), with five of these residues having a mutational frequency higher than 1σ (residues 60, 62, 63, 64, and 65). Residue 76 (in blue) corresponds to the substitution that restores WT-β2m. Residue 66 was the only residue with a mutational frequency lower than 1σ to have a positive effect on D76N-β2m. (B) In vivo growth scores of 15 variants (D76N + X) compared with WT-β2m and D76N-β2m. In total, 13 variants were found in the APR (A, in green), and two variants were selected using Solubis (in orange; SI Appendix, Fig. S12). Data represent the mean (n = three biologically independent repeats), where each point corresponds to one experiment. The variants are ordered from the highest to lowest in vivo growth score (AUC) (Left to Right). The error bar represents one SD. Note that a larger range of ampicillin concentration was used to determine the behavior of these improved variants (0 to 280 μg mL−1) (SI Appendix, Materials and Methods); hence, the AUC is greater than those shown for D76N-β2m and WT-β2m in Figs. 1D and 3. (C) Protein stability (Tm;app) for WT-β2m, D76N-β2m, and six variants containing amino acid changes in the APR (i.e., double variants D76N + X, where X is a residue in the APR) obtained using temperature ramp monitored by far-UV CD. The error bar is the fitting error. (D) Protein aggregation rate (Thalf) for WT-β2m, D76N-β2m, and four of the double variants, determined using ThT fluorescence. The error bar represents one SD of 8 to 10 repeats. Note that insufficient D76N_L64P- and D76N_L65P-β2m could be purified to perform the aggregation assay. Asterisks denote significance (t test: paired two sample for means, two-tail) where *P = 0.04 and ***P < 0.0001. Note that the same color code is used for B–D.