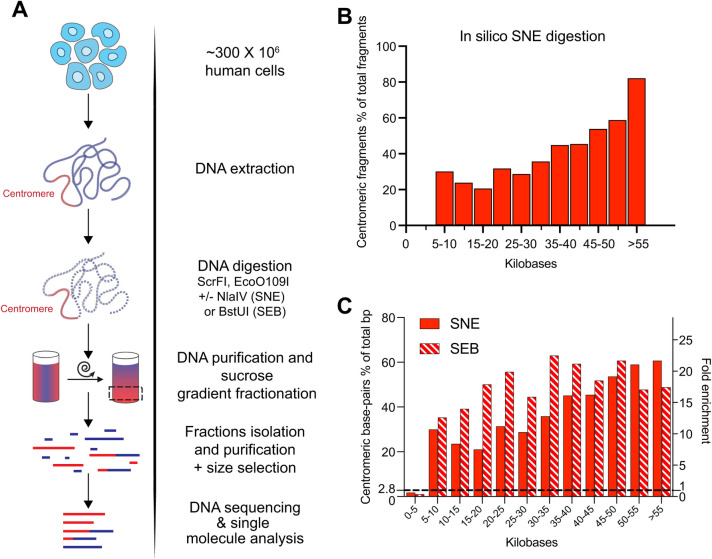
Fig 1. A restriction enzyme-based method to enrich and purify centromeric DNA from human cells.
A. Schematic representation of the experimental design. B. Predicted distribution of the percentage of centromeric fragments in the indicated size bins after in silico digestion of the reference T2T-CHM13v1.0 genome with the SNE enzyme combination. Y-axis represents the percentage of centromeric fragments over total fragments in each length range. C. Distribution of centromeric base-pairs according to predicted fragment length after in silico digestion of the reference T2T-CHM13v1.0 genome with the SNE or SEB enzyme combinations. The y-axis on the left represents the percentage of centromeric base-pairs over total base-pairs in each length range. The dotted line at 2.8% represents the percentage of centromeric base-pairs in the reference genome, corresponding to the expected fraction of centromeric DNA in a theoretical non-enriched sample. The y-axis on the right reports the fold enrichment in centromeric base-pairs over the non-enriched sample (~2.8% of centromeric base-pairs in the reference genome).