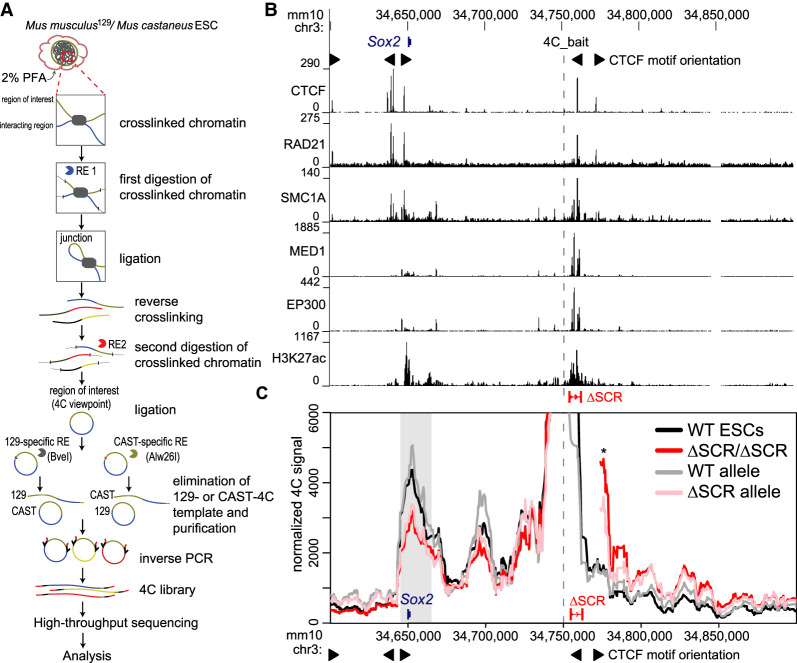
Figure 1.
Deletion of the SCR partially disturbs chromatin interactions with the Sox2 gene in ESCs. (A) Schematic of the allele-specific 4C approach. (RE) Restriction enzyme, (PFA) paraformaldehyde. (B) The region surrounding the Sox2 gene is displayed on the UCSC genome browser (mm10). The SCR deletion (ΔSCR) is shown, and the 4C bait region is indicated as a dashed line. ChIP-seq conducted in ESCs is shown below for CTCF, RAD21, SMC1A, MED1, EP300, and H3K27ac. The motif orientations of bound CTCF sites are denoted. (C) 4C data are shown for wild-type cells (WT, black, n = 4), homozygous ΔSCR/ΔSCR cells (red, n = 4), and heterozygous ΔSCR cells. Data from the heterozygous cells are displayed separately for the WT (gray, n = 3) and ΔSCR (pink, n = 4) alleles. The dashed line indicates the location of the 4C bait region. The gray box indicates the bait-interacting region surrounding the Sox2 gene. Compared with WT cells, a significant decrease in relative interaction frequency of the 4C bait region with the Sox2 gene was observed for homozygous ΔSCR/ΔSCR cells (P = 0.02) and the ΔSCR allele in heterozygous ΔSCR cells (P = 0.04), but not the WT allele in heterozygous ΔSCR cells (P = 0.46). An asterisk denotes the region now contiguous with the bait in ΔSCR alleles, explaining the very high 4C signal. For deletion alleles, the 4C signal has been omitted from the deleted region and flanking positions that are also affected by the deletion when computing running means. The motif orientations of bound CTCF sites are denoted.