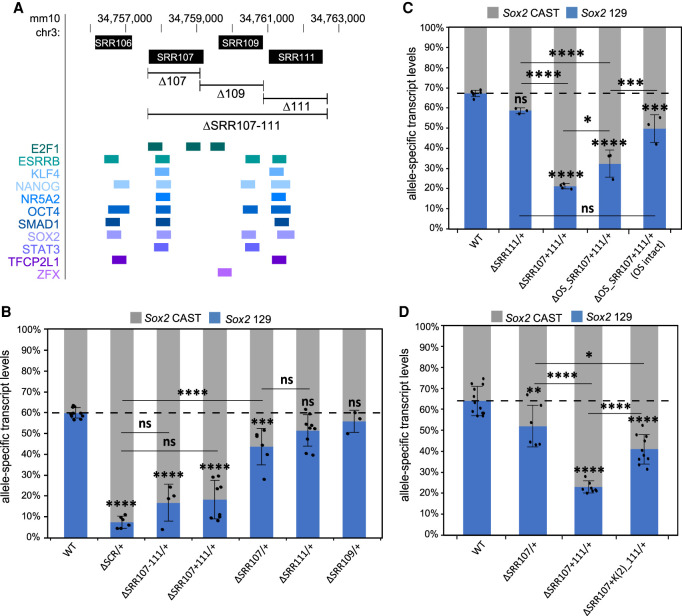
Figure 2.
SRR107 and SRR111 have a partially redundant role and are required for the transcription-enhancing capacity of the SCR. (A) The SCR genomic region is displayed on the UCSC genome browser (mm10). Sox2 regulatory regions (SRRs, top) correspond to transcription factor-bound regions derived from ESC ChIP-seq data sets compiled in the CODEX database (bottom). In B–D, allele-specific primers detect Sox2 musculus (129; blue) or castaneus (CAST; gray) mRNA by RT-qPCR from F1 ESC clones from the indicated genotype. Expression levels for each allele are shown relative to the total transcript levels. Error bars represent SD. n ≥ 3. Significant differences from the WT values are indicated. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001, (ns) not significant. (B) Deletion of both SRR107 and SRR111 (ΔSRR107–111/+; ΔSRR107+111/+) causes a reduction in Sox2 transcript levels similar to that for deletion of the entire SCR. (C) Deletion of the OCT4:SOX2 (OS) motif in SRR107 (ΔOS_SRR107+111/+) reduces Sox2 transcript levels on the linked allele. Clones with nucleotide deletions near but not disrupting the OS motif (ΔOS_SRR107+111/+ [OS intact]) displayed increased transcription of Sox2 on the linked allele compared with clones with a deleted OS motif. (D) Deletion of two KLF4 motifs in SRR111 [ΔSRR107+K(2)_111/+] reduces Sox2 transcript levels on the linked allele.