Abstract
Immunoglobulins (Igs) are major serum proteins which play important roles in immunity. Both untargeted and targeted proteomic workflows can be applied to investigate antigen-binding sites and the glycosylation profiles of Igs. For a more-comprehensive picture of IgG from human serum, we developed an IgG purification process and coupled the standardized method to untargeted and targeted proteomic workflows for IgG investigations. Parameters such as the type of purification beads, volume of the bead slurry, incubation conditions, and binding capacities were evaluated in this study. Only 2 μL of human serum was required for each sample. The performance of coupling the purification process to untargeted proteomics in the IgG analysis was evaluated by comparing normalized abundances of IgG subclass-specific peptides with quantification results from an ELISA. Pearson’s correlation values were all >0.82. Targeted proteomic workflow was applied to serum samples from patients with autoimmune pancreatitis and from healthy controls, and the results corresponded to clinical findings that IgG4-related peptides/glycopeptides showed higher abundances in the diseased group. The developed IgG purification process is simple and requires small sample volume, and it can be coupled to targeted and untargeted proteomic workflows for clinical investigations in the future.
Keywords: IgG purification, Proteomics, Liquid chromatography-mass spectrometry, Human serum
1. Introduction
Proteomics-based study is one of the approaches for investigating the antibody repertoire. Recently, many untargeted proteomics studies investigated immunoglobulin G (Ig; IgG) or specific autoantibodies which were purified with affinity columns or antigen-immobilizing beads using liquid chromatography-high-resolution mass spectrometry (LC-HRMS). MS/MS spectra were used to identify peptide sequences, and differences in peptides from Ig-variable regions were compared among groups or for patients with a specific disease. Luider et al. applied a proteomics-based approach to an IgG-variable region analysis, and results showed that special variable-region peptides with mutated sequences were shared among patients with multiple sclerosis [1]. For cancer diagnoses, they also identified 12 specific IgG variable-region peptides which can be used to generate a regression model for distinguishing between patients with lung cancer and healthy controls [2]. The model was able to differentiate patients with lung cancer from healthy controls with high sensitivity (84%) and specificity (90%). Kurtin et al. applied a proteomics-based antibody study to analyze light-chain peptides from biopsies of patients with systemic Ig light-chain amyloidosis (AL). From their studies, some light-chain peptides were only detected in AL patients, and expressions of these light-chain peptides were correlated with organ failure and the degree of protein precipitation [3]. Gordon et al. investigated autoantibodies from patients with systemic lupus erythematosus. In their study, anti-Smith autoantibodies were purified from the serum of six patients, and variable-region peptides were investigated. Results showed that the six patients shared the same Ig germline, and the same mutated sequences were shared among the six patients [4]. Schmelter et al. applied a proteomics-based approach to primary open-angle glaucoma, and 75 peptides from the Ig-variable region were only found in patients [5]. Findings of antibody variable-region sequences have the potential to be used for disease diagnosis, and provide novel information about pathophysiology.
Instead of analyzing the whole antibody, subunits of Igs are usually fractionated before an untargeted proteomics analysis. Heavy-chain and light-chain subunits can be separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and an in-gel digestion process is applied before a liquid chromatographic mass spectrometric (LC-MS) analysis [1,4,6,7]. On the other hand, Fab of immunoglobulins can also be separated from the Fc region by SDS-PAGE or by an on-bead digestion process, and the Fab fraction was introduced for investigations [2,5,8–10]. Fab analyses are relatively efficient in terms of simultaneously collecting peptide information from heavy-chain and light-chain variable regions; however, it can be difficult to identify whether the peptide of interest is from a heavy chain or light chain. For IgG purification, most studies use spin columns packed with Melon gel resin or an anti-IgG-Fc affinity matrix to purify IgG from serum or cerebrospinal fluid (CSF) [1,2,5,8–10]. In those studies, 80–200 μL of serum or CSF was diluted and introduced to spin columns for incubation. In addition, Dekker at el. used a fast protein LC (FPLC) system coupled with a protein G affinity column to purify IgG from clinical samples, for which 2mL of serum was used [10]. The eluate was neutralized and concentrated with an ultrafiltration unit.
Targeted proteomic studies have also been applied to many IgG studies. LC coupled to tandem MS (LC-MS/MS) was used to quantify concentrations of IgG subclasses [11–13]. Gugten et al. found that quantification results of IgG subclasses from immunonephelometry, the most commonly used method for clinical samples, had a bias regarding IgG2. In contrast, LC-MS/MS provided more-precise results when determining IgG subclasses [12]. By utilizing multiple-reaction monitoring detection, glycopeptides of the IgG-Fc region can also be monitored [14]. Many studies showed that glycosylation profiles of IgG were correlated with different diseases, such as colorectal cancer, breast cancer, and inflammatory bowel disease [15–17]. Recently, Konno et al. also found that N-glycans of IgG4 from patients with IgG4-related disease differed from those purified from healthy controls [18]. Kazuno et al. also correlated the IgG glycosylation profile with the status of prostate disease [19]. Direct serum protein digestion or IgG purification followed by protein digestion was performed in those studies, and the serum volume used was generally 2–20 μL.
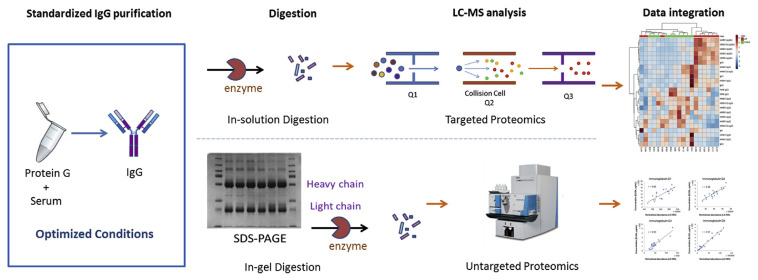
In order to obtain a more-comprehensive profile of IgG from clinical samples, we wanted to analyze peptides from variable regions and the IgG-Fc glycan using both untargeted and targeted proteomic workflows (Fig. 1). A standardized IgG purification process was developed and only 2 μL of serum was required for IgG purification. Performances of coupling the purification process to untargeted and targeted IgG proteomics studies were also investigated in this report.
Fig. 1.
Workflows of the targeted and untargeted proteomic studies for immunoglobulin G (IgG) analyses. The standardized IgG purification process can be coupled to both workflows in clinical applications.
2. Methods
2.1. Standard and reagents
Protein G Mag SepharoseXtra™ was purchased from GE Healthcare (Uppsala, Sweden). Polymerase chain reaction (PCR) tubes (0.2 mL) were purchased from Labcon (Petaluma, CA, USA). An enzyme-linked immunosorbent assay (ELISA) kit for the Human IgG Subclass Profile was purchased from Life Technologies (product # 991,000, Rockville, MD, USA). LC-grade acetonitrile (ACN) was purchased from J.T. Baker, Avantor Performance Materials (Center Valley, PA, USA). The magnetic stand (DynaMagTM-96 Side) for the PCR tubes was purchased from Life Technologies (Oslo, Norway). Trifluoroacetic acid (TFA) and formic acid were purchased from Sigma Aldrich (St. Louis, MO, USA). Trypsin was purchased from Promega (Madison, WI, USA).
2.2. Purification of serum IgG with magnetic beads
The IgG purification process was performed in 0.2-mL PCR tubes. Twenty microliters of protein G magnetic bead slurry were added to pre-aliquoted 150 μL of phosphate-buffered saline (PBS) binding buffer (pH 7.4) for equilibration. After equilibration, the buffer and slurry were removed, and another 150 μL of fresh PBS binding buffer was added to suspend the magnetic beads. Two microliters of human serum was spiked into the binding buffer, and the sample was vortex-mixed. Sample incubation was set up with end-to-end shaking on a desktop shaker (ELMI, Intelli-Mixer RM-2M, Riga, LV-1006, Latvia), and the temperature was kept at 4 °C overnight. The binding buffer was then removed, and the beads were first washed with 150 μL of PBS buffer and then with 150 μL of deionized water. IgG was finally eluted using 80 μL of 100 mM formic acid (pH 2.9) twice. Two eluents were combined for drying with a centrifugal evaporator (LaboGene, Allerød, Denmark), and 2 μL of eluent was used for IgG quantification using a NanoDrop spectrophotometer (Thermo Fisher Scientific).
2.3. ELISA experiments for total IgG and IgG subclass quantification
ELISAs were used to determine concentrations of total IgG and IgG subclasses from control (untreated) and purified IgG samples. The total IgG ELISA experiment was used to investigate the purification parameters and purification recovery. The IgG subclass ELISA assay was used to validate the workflow from IgG purification to untargeted proteomics analysis by comparing ELISA quantification results with normalized abundances of IgG subclass-specific peptides calculated by Progenesis QI software based on LC-MS/MS results, where log space of ratio abundance of peptide ions were used to estimate the scalar factor and applied to each sample run [20]. Control serum samples were diluted with ELISA diluent buffer for the assay. Purified IgG samples were reconstituted with ELISA diluent buffer, and were further diluted to theoretical concentrations before the analysis. The ELISA processes were performed according to the manufacturer’s protocol. The data analysis and quantification were performed using Microsoft Excel 2010 and GraphPad Prism 5.
2.4. In-gel digestion for IgG heavy chain
According to the quantification results from the NanoDrop spectrophotometer, each sample was loaded onto a 10% polyacrylamide gel (10 × 10 cm, 0.75 mm thick) with a protein concentration of 8 μgmL−1 for reducing SDS-PAGE separation. The heavy chain and light chain of IgG were observed using Coomassie blue staining. The lane of the IgG heavy chain was collected and nebulized into gel pieces with a gel nebulizer. The gel pieces were washed with deionized water for 10 min twice and de-stained using 25 mM NH4HCO3/50% ACN. The washed gel pieces were treated with 10 mM DTT and 55 mM iodoacetamide (IAA) in a 25 mM NH4HCO3 solution for reduction and alkylation, respectively. Protein digestion was performed at 37 °C overnight with the addition of modified trypsin (Promega). Digested peptides were extracted with 1% TFA/60% ACN, and dried using a centrifugal evaporator. The sample was reconstituted with 40 μL of 0.1% formic acid for the LC-MS/MS untargeted proteomics analysis.
2.5. In-solution digestion for purified IgG samples
The in-solution digestion process was chosen to treat samples for the targeted proteomics analysis. Purified IgG samples were reconstituted with 50 μL of 50 mM NH4HCO3. One microliter of dithiothreitol (DTT) which was freshly prepared in water (550 mM) was added to a sample for disulfide bond reduction. Samples were kept at 56 °C for 45 min. Two microliters of IAA (450 mM) was added to the samples, and samples were kept at room temperature for another 45 min. Trypsin was used for protein digestion, and 1 μg of trypsin was added to each sample. Samples were kept at 37 °C for 16–18 h.
2.6. LC-MS/MS analysis for the untargeted and targeted proteomics studies
For untargeted proteomics, an LC analysis was performed using an Agilent 1100 HPLC system equipped with a binary solvent pump (Agilent Technologies, Palo Alto, CA, USA), and a FAMOS autosampler with the sample reservoir maintained at 4 °C (LC Packings, CA, USA). The trapping column (150 μm I.D. × 15 mm) and separation column (75 μm I.D. × 300 mm) were packed with C18 beads (5 μm, 100 Ä) for the reversephase LC analysis. A sample was injected into the trapping column at a volume of 5 μL and a flow rate of 10 μLmin−1. The mobile phase was composed of 0.1% formic acid in water (solvent A) and 0.1% formic acid in 80% ACN (solvent B) at a flow rate of 300 nL min−1. The mobile phase was maintained at 2% solvent B for 2.5 min during sample trapping, which was increased to 10% solvent B in 1 min. The linear gradient was then increased from 10% to 40% solvent B from 3.5 to 42 min, and increased to 98% solvent B in the following 4 min. The percentage was kept at 98% solvent B for 4 min, and the system was re-equilibrated to 2% solvent B for 10 min. The MS analysis was performed using a 7-T LTQ-FT-ICR (linear trap quadrupole-Fourier transformation-ion cyclotron resonance) mass spectrometer (Thermo Fisher Scientific, MA, USA) coupled with a nanoelectrospray ion source (New Objective, MA, USA). A positive electrospray ionization mode was used with the following parameters: a source voltage of 1.4 kV, a capillary voltage of 9 V, and a capillary temperature of 200 °C. Full-scan MS spectra ranging m/z 320–2000 were acquired by the FT-ICR MS with a resolution of 100,000 at m/z 400. The five most abundant ions from the full-scan MS spectrum were selected for the MS/MS analysis. The normalized collision energy of CID was set to 35. LC-MS data were processed and analyzed by using Progenesis QI software.
For targeted proteomics, the analysis utilized an Agilent 1260 high-performance LC (HPLC) system equipped with an Agilent 6470 triple quadruple system (Agilent Technologies, Waldbronn, Germany). The separation LC column was Core-Shell C18 Kinetix® (50 × 2.1 mm, 2.6 μm) with a mobile phase consisting of 0.1% aqueous formic acid (solvent A) and 0.1% formic acid in ACN (solvent B) at a flow rate of 0.4 mL min−1. The injection volume was 5 μL. The following was the gradient: mobile phase at 5% B for 0.5 min, and the organic phase was increased to 50% B (50% A) in 9.5 min. The organic phase was then changed to 100% in 1min with a linear gradient, and restored to 5% B (95% A) in 1min at a flow rate of 0.6 mL min−1 and maintained for 3 min. The column was re-equilibrated with 5% B (0.4 mL/min) for 2 min. The sample reservoir was set to 8 °C, while the column oven was set to 40 °C. The positive electrospray ionizationmode was selected. The source parameters were as follows: a gas temperature of 300 °C; gas flow of 5 L/min; nebulizer at 35 psi; sheath gas heater set to 325 °C; sheath gas flow of 11 L/min; and capillary voltage of 3500 V. The transitions for selected peptides/glycopeptides are listed in Table 1 [14]. Serum samples from healthy volunteer were used to evaluate the repeatability of peak area. Three batches of samples were prepared and analyzed on two occasions. The overall analytical reproducibility (RSD, %) in terms of peak area for peptides and glycopeptides were less than 15.1% and 24.2%, respectively. H3N5F1-IgG1 and H4N4F1S1-IgG1 have RSD higher than 25% due to the smaller signal intensities.
Table 1.
Parameters of multiple reaction monitoring (MRM) for targeted peptide/glycopeptide analysis.
| Compound name | FVd (V) | CEe (V) | Precursor ion (m/z) | Product ion (m/z) |
|---|---|---|---|---|
| H3N4F1-IgG1a | 135 | 15 | 878.8 | 204.1 |
| H3N4F1-IgG2b | 135 | 15 | 868.1 | 204.1 |
| H3N4F1-IgG3/4c | 135 | 15 | 873.4 | 204.1 |
| H3N5F1-IgG1 | 135 | 15 | 946.5 | 204.1 |
| H3N5F1-IgG2 | 135 | 15 | 935.8 | 204.1 |
| H3N5F1-IgG3/4 | 135 | 15 | 941.1 | 204.1 |
| H4N4F1-IgG1 | 135 | 15 | 932.8 | 204.1 |
| H4N4F1-IgG2 | 135 | 15 | 922.1 | 204.1 |
| H4N4F1-IgG3/4 | 135 | 15 | 927.4 | 204.1 |
| H4N4F1S1-IgG1 | 135 | 15 | 1029.8 | 204.1 |
| H4N4F1S1-IgG2 | 135 | 15 | 1019.1 | 204.1 |
| H4N4-IgG1 | 135 | 15 | 884.1 | 204.1 |
| H4N4-IgG2 | 135 | 15 | 873.4 | 204.1 |
| H4N5F1-IgG1 | 135 | 15 | 1000.5 | 204.1 |
| H4N5F1-IgG2 | 135 | 15 | 989.9 | 204.1 |
| H5N4F1-IgG2 | 135 | 15 | 976.1 | 366.1 |
| H4N5F1-IgG3/4 | 135 | 15 | 995.1 | 204.1 |
| H4N5-IgG1 | 135 | 15 | 951.7 | 204.1 |
| H4N5-IgG2 | 135 | 15 | 941.1 | 204.1 |
| H5N4F1-IgG1 | 135 | 15 | 986.8 | 366.1 |
| H5N4F1S1-IgG1 | 135 | 15 | 1083.8 | 366.1 |
| H5N4F1S1-IgG2 | 135 | 15 | 1073.1 | 366.1 |
| H5N4F1S1-IgG3/4 | 135 | 15 | 1078.4 | 366.1 |
| H5N4-IgG1 | 135 | 15 | 938.1 | 366.1 |
| H5N5F1-IgG1 | 135 | 15 | 1054.5 | 366.1 |
| H5N5F1-IgG2 | 135 | 15 | 1043.8 | 366.1 |
| IgG (DTLMISR) | 135 | 9 | 418.2 | 619.4 |
| 9 | 418.2 | 506.3 | ||
| IgG1 (GPSVFPLAPSSK) | 135 | 20 | 593.8 | 846.5 |
| 20 | 593.8 | 699.4 | ||
| IgG2 (GLPAPIEK) | 135 | 15 | 412.7 | 654.4 |
| 15 | 412.7 | 486.3 | ||
| IgG3 (WYVDGVEVHNAK) | 135 | 6 | 472.9 | 697.4 |
| 6 | 472.9 | 534.3 | ||
| IgG4 (TTPPVLDSDGSFFLYSR) | 135 | 9 | 635.0 | 425.2 |
| 15 | 951.6 | 850.6 |
IgG1 glycopeptide: glycan-EEQYNSTYR; H, hexose; N, N-acetylglucosamine; F, fucose; S, N-acetylneuraminic acid.
IgG2 glycopeptide: glycan-EEQFNSTFR.
IgG3/4 glycopeptide: glycan-EEQYNSTFR (IgG3); glycan-EEQFNSTYR (IgG4).
FV: fragmentor voltage.
CE: collision energy.
2.7. Clinical samples
The developed IgG purification method was coupled to targeted and untargeted proteomics workflows, which were applied to clinical samples. Serum samples were collected from patients with a diagnosis of autoimmune pancreatitis (AIP, n = 10) and from healthy volunteers (n = 10) for both targeted and untargeted proteomics workflows, respectively. This study was approved by the research ethics committee of National Taiwan University Hospital (201301048RIND), and signed informed consent forms were received from all patients and participants who joined the study.
3. Results and discussion
In this study, we chose protein G-immobilized material to purify all four IgG subclasses from human serum. Compared to protein G Sepharose beads and packed affinity columns, magnetic beads provide convenience in manipulation in that magnetic beads can efficiently be separated from the incubation and wash solutions using a magnetic stand. Therefore, protein G magnetic beads with different specifications were selected for comparison in this study. The volume of the bead slurry, incubation time, and capacity of magnetic beads for IgG purification were optimized and investigated.
3.1. Comparison of IgG purification beads
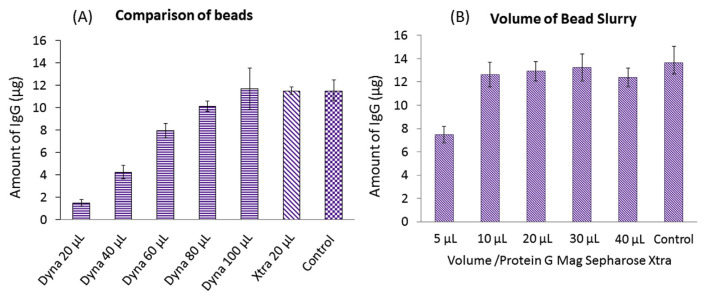
With the convenience of using magnetic beads, two different brands of protein G immobilized beads were compared. Dynabeads™ Protein G (Thermo Fisher Scientific) are composed of 2.8-μm magnetic beads which covalently couple with recombinant protein G. The particle size of Protein GMag SepharoseXtra (GE) ranges 37–100 μm. Their respective purification capacities are 8 and 27 μg human IgG/μL magnetic beads. Two microliters of human serum was used to optimize volumes of the bead slurry, which were needed to achieve the best purification efficiency. By comparing the IgG amount with control samples with no purification, 100 μL of Dynabeads™ Protein G was needed to purify IgG from 2 μL of serum (Fig. 2A). Using a similar optimization approach, 20 μL of Protein G Mag SepharoseXtra was sufficient to purify IgG from 2 μL of serum (Fig. 2B). With benefits of smaller volumes of the bead slurry required, faster separation of beads from solution (with a large size of magnetic beads), and cost savings, 20 μL of Protein G Mag SepharoseXtra was chosen for IgG purification.
Fig. 2.
Optimization of the volumes of the bead slurry for two different brands of protein G magnetic beads. (A) Optimization results for Dynabeads protein G magnetic beads and comparison with 20 μL of Protein G Mag SepharoseXtra. (B) Optimization results for Protein G Mag SepharoseXtra.
3.2. Optimization of temperature and time for IgG purification
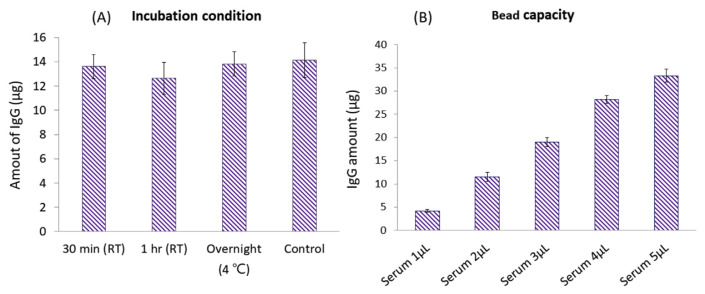
After optimizing volumes of the bead slurry for IgG purification from 2 μL of serum, we also investigated the time and temperature of sample incubation. With the use of 20 μL protein G magnetic beads, we compared the incubation process at room temperature for 0.5 and 1 h, and at 4 °C overnight. All three conditions showed good extraction efficiencies compared to control serum samples directly quantified by an ELISA (Fig. 3A). To ensure if 20 μL of protein G magnetic beads was sufficient for future applications if different clinical samples contained higher or lower amounts of IgG, we spiked different volumes of human serum (1–5 μL) in PBS to mimic different concentrations of IgG in samples. With the optimized purification process, samples with IgG amounts ranging 4–35 μL could be purified and quantified by an ELISA (Fig. 3B). Results showed that 20 μL of selected protein G beads provided high efficiency of IgG purification in which a 30-min incubation at room temperature was sufficient to achieve high recovery. In addition, 20 μL of beads showed a high binding capacity which can purify higher concentrations of IgG from clinical samples in future applications.
Fig. 3.
Optimization of incubation temperature and time for immunoglobulin G (IgG) binding (A), and investigation of the bead-binding capacity of 20 μL of protein G magnetic beads (B).
3.3. Application of the IgG purification process to untargeted proteomics antibody investigations
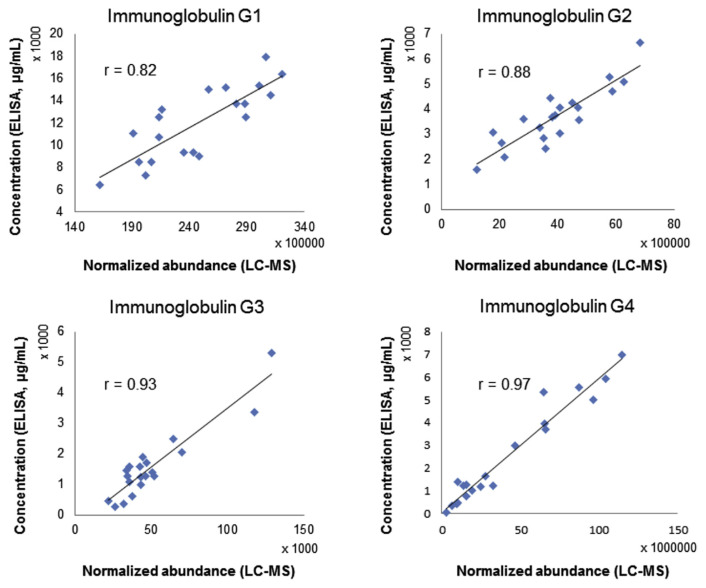
In order to evaluate if the IgG purification process can be coupled to untargeted proteomics studies and be applied for antibody repertoire studies, we applied the workflow to clinical samples and compared the normalized responses of IgG subclass-specific peptides to IgG ELISA quantification results. Peptides which represent IgG1 to IgG4 are GPSVFPLAPSSK (m/z 593.8269), GLPAPIEK (m/z 412.7471), WYVDGVEVHNAK (m/z 472.9017), TTPPVLDSDGSFFLYSR (m/z 951.4676). Pearson’s correlation values were all >0.82 (Fig. 4). Although the verification method of using an ELISA in parallel cannot reflect the accuracy of relative quantification for peptides from IgG-variable regions, it is still an early checkpoint method to ensure that the workflow caused no significant bias in relative quantification.
Fig. 4.
Correlation of normalized abundances of immunoglobulin G (IgG) subclass-specific peptides from untargeted proteomic workflow to quantification results using an ELISA.
3.4. Application of the IgG purification process to targeted proteomics Ig investigations
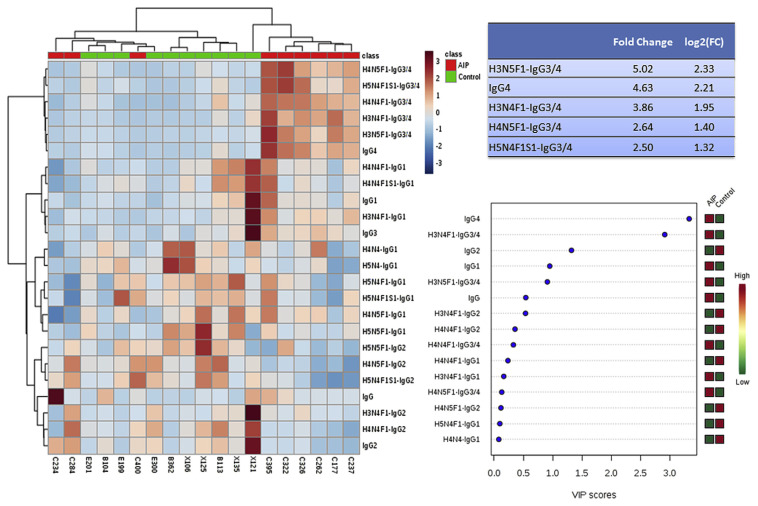
We also coupled the developed IgG purification process to target proteomics workflow, where in-solution digestion was utilized before sample injection. IgG-specific peptides and IgG-Fc glycopeptides were detected using LC-MS/MS in the multiple reaction monitoring (MRM) mode. We purified IgG from ten healthy volunteers and ten patients with AIP. After evaluating the technical repeatability of peak area, 24 targets with RSDs less than 15% among at least 85% of samples were selected. The peak area of each target was merged into a single matrix, and profiles of IgG glycopeptides were analyzed using a multivariate analysis. One data set from patient with auto-immune pancreatitis was found to be outlier in principal component analysis, and was excluded before further data analysis. From results of Fig. 5, one can see that some of the IgG3/4-related glycopeptides and IgG4 showed higher quantities in serum from AIP patients. This finding corresponds to clinical symptoms that AIP is an IgG4-related disease, and quantities of IgG4 or total IgG are usually high in patient sera. Three of the AIP patients were classified as healthy. Two of them showed similar IgG glycopeptide profiles as healthy individuals, and one patient had high total IgG and IgG2 profiles. From the application, we also found that IgG1 and IgG3 were relatively higher in the AIP group, while IgG2 was relatively lower. It is worth applying the developed IgG purification process and targeted proteomics workflow to more clinical samples and IgG studies in the future.
Fig. 5.
Application of targeted proteomic workflow to clinical samples from patients with autoimmune pancreatitis and from healthy individuals. A heatmap was generated using 24 targets, and the fold changes for the top five targets were calculated. Variable’s importance (VIP) scores were calculated from a partial least squares discrimination analysis.
3.5. Potential of current workflow in IgG investigation
Compared with previous literature that using untargeted proteomics in IgG variable region studies (serum volume > 80 μL), the current method of IgG purification could be achieved by using 2 μL of serum only [2,5,8–10]. In addition, the purification process can be used in targeted proteomics for IgG-Fc glycopeptides and IgG subclasses investigation. Dr. Mattman and Dr. Murray et al. have developed LC-MS/MS methods to determine IgG subclasses from serum, and 20 μL of serum samples were used without IgG purification [11,12]. Dr. Pan et al. also applied LC-MS/MS to IgG subclasses investigation, and 25 μL of plasma samples were used in the study [13]. Our current method is much more sample saving. Dr. Ruhaak et al. also used 2 μL of serum samples and direct in solution protein digestion to quantify IgG and glycoforms by using LC-MS/MS [14]. Although our current method included IgG purification which is relatively time consuming, the developed purification process can be coupled to untargeted proteomics IgG variable region investigation where purification is necessary. In addition, it could decrease matrix effects during MS detection for targeted proteomics.
4. Conclusions
For the antibody repertoire study, targeted and untargeted proteomic approaches can be used to investigate Ig profiles and special antigen-binding sites. In this study, we optimized and developed an IgG purification process that only required 2 μL of human serum or plasma for each clinical sample. We coupled the purification process with in-gel digestion and untargeted proteomics, and normalized abundances of IgG subclass-specific peptides showed good correlations with ELISA results. We also coupled the purification process to insolution digestion and the targeted proteomics method to analyze IgG profiles of AIP patients and healthy controls. The results corresponded to AIP clinical symptoms, which means that the developed purification process is capable of being applied to IgG-related studies. With the developed IgG purification method, both untargeted and targeted IgG investigation methods can be utilized in clinical research in the future.
Acknowledgements
This study was supported by the Ministry of Science and Technology (105-2320-B-038-066-; 106-2320-B-038-050-MY2). We thank the technical support provided by TMU Core Facility. We would like to acknowledge Mr. Chun-Chih Jared Liu and Ms. Yuan-Chin Hsiung for their excellent technical support at TMU Core Facility.
Funding Statement
This study was supported by the Ministry of Science and Technology (105-2320-B-038-066-; 106-2320-B-038-050-MY2).
References
- 1. Singh V, Stoop MP, Stingl C, Luitwieler RL, Dekker LJ, van Duijn MM, et al. Cerebrospinal-fluid-derived immunoglobulin G of different multiple sclerosis patients shares mutated sequences in complementarity determining regions. Mol Cell Proteomics MCP. 2013;12:3924–34. doi: 10.1074/mcp.M113.030346. [In eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Costa D, Broodman I, Calame W, Stingl C, Dekker LJ, Vernhout RM, et al. Peptides from the variable region of specific antibodies are shared among lung cancer patients. PLoS One. 2014;9:e96029. doi: 10.1371/journal.pone.0096029. [In eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dasari S, Theis JD, Vrana JA, Meureta OM, Quint PS, Muppa P, et al. Proteomic detection of immunoglobulin light chain variable region peptides from amyloidosis patient biopsies. J Proteome Res. 2015;14:1957–67. doi: 10.1021/acs.jproteome.5b00015. [In eng] [DOI] [PubMed] [Google Scholar]
- 4. Al Kindi MA, Chataway TK, Gilada GA, Jackson MW, Goldblatt FM, Walker JG, et al. Serum SmD autoantibody proteomes are clonally restricted and share variable-region peptides. J Autoimmun. 2015;57:77–81. doi: 10.1016/j.jaut.2014.12.005. [In eng] [DOI] [PubMed] [Google Scholar]
- 5. Schmelter C, Perumal N, Funke S, Bell K, Pfeiffer N, Grus FH. Peptides of the variable IgG domain as potential biomarker candidates in primary open-angle glaucoma (POAG) Hum Mol Genet. 2017;26:4451–64. doi: 10.1093/hmg/ddx332. [In eng] [DOI] [PubMed] [Google Scholar]
- 6. Al Kindi MA, Colella AD, Beroukas D, Chataway TK, Gordon TP. Lupus anti-ribosomal P autoantibody proteomes express convergent biclonal signatures. Clin Exp Immunol. 2016;184:29–35. doi: 10.1111/cei.12750. [In eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dekker LJ, Zeneyedpour L, Brouwer E, van Duijn MM, Sillevis Smitt PA, Luider TM. An antibody-based biomarker discovery method by mass spectrometry sequencing of complementarity determining regions. Anal Bioanal Chem. 2011;399:1081–91. doi: 10.1007/s00216-010-4361-9. [In eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Costa D, Broodman I, Vanduijn MM, Stingl C, Dekker LJ, Burgers PC, et al. Sequencing and quantifying IgG fragments and antigen-binding regions by mass spectrometry. J Proteome Res. 2010;9:2937–45. doi: 10.1021/pr901114w. [In eng] [DOI] [PubMed] [Google Scholar]
- 9. Broodman I, de Costa D, Stingl C, Dekker LJ, VanDuijn MM, Lindemans J, et al. Mass spectrometry analyses of kappa and lambda fractions result in increased number of complementarity-determining region identifications. Proteomics. 2012;12:183–91. doi: 10.1002/pmic.201100244. [In eng] [DOI] [PubMed] [Google Scholar]
- 10. Dekker L, Wu S, Vanduijn M, Tolic N, Stingl C, Zhao R, et al. An integrated top-down and bottom-up proteomic approach to characterize the antigen-binding fragment of antibodies. Proteomics. 2014;14:1239–48. doi: 10.1002/pmic.201300366. [In eng] [DOI] [PubMed] [Google Scholar]
- 11. Ladwig PM, Barnidge DR, Snyder MR, Katzmann JA, Murray DL. Quantification of serum IgG subclasses by use of subclass-specific tryptic peptides and liquid chromatography–tandem mass spectrometry. Clin Chem. 2014;60:1080–8. doi: 10.1373/clinchem.2014.222208. [In eng] [DOI] [PubMed] [Google Scholar]
- 12. van der Gugten G, DeMarco ML, Chen LYC, Chin A, Carruthers M, Holmes DT, et al. Resolution of spurious immunonephelometric IgG subclass measurement discrepancies by LC-MS/ MS ClinChem. 2018;64:735–42. doi: 10.1373/clinchem.2017.282319. [Ineng] [DOI] [PubMed] [Google Scholar]
- 13. ZHXDP Identification and quantification of immunoglobulin G (G1, G2, G3 and G4) in human blood plasma by high-resolution quadrupole-Orbitrap mass spectrometry. RSC Adv. 2017;7:20212–8. [Google Scholar]
- 14. Hong Q, Lebrilla CB, Miyamoto S, Ruhaak LR. Absolute quantitation of immunoglobulin G and its glycoforms using multiple reaction monitoring. Anal Chem. 2013;85:8585–93. doi: 10.1021/ac4009995. [In eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawaguchi-Sakita N, Kaneshiro-Nakagawa K, Kawashima M, Sugimoto M, Tokiwa M, Suzuki E, et al. Serum immunoglobulin G Fc region N-glycosylation profiling by matrix-assisted laser desorption/ionization mass spectrometry can distinguish breast cancer patients from cancer-free controls. Biochem Biophys Res Commun. 2016;469:1140–5. doi: 10.1016/j.bbrc.2015.12.114. [In eng] [DOI] [PubMed] [Google Scholar]
- 16. Vuckovic F, Theodoratou E, Thaci K, Timofeeva M, Vojta A, Stambuk J, et al. IgG glycome in colorectal cancer. Clin Cancer Res – Offic J Am Assoc Cancer Res. 2016;22:3078–86. doi: 10.1158/1078-0432.CCR-15-1867. [In eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trbojevic Akmacic I, Ventham NT, Theodoratou E, Vuckovic F, Kennedy NA, Kristic J, et al. Inflammatory bowel disease associates with proinflammatory potential of the immunoglobulin G glycome. Inflamm Bowel Dis. 2015;21:1237–47. doi: 10.1097/MIB.0000000000000372. [In eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Konno N, Sugimoto M, Takagi T, Furuya M, Asano T, Sato S, et al. Changes in N-glycans of IgG4 and its relationship with the existence of hypocomplementemia and individual organ involvement in patients with IgG4-related disease. PLoS One. 2018;13:e0196163. doi: 10.1371/journal.pone.0196163. [In eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kazuno S, Furukawa J, Shinohara Y, Murayama K, Fujime M, Ueno T, et al. Glycosylation status of serum immunoglobulin G in patients with prostate diseases. Cancer Med. 2016;5:1137–46. doi: 10.1002/cam4.662. [In eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Progenesis QI for proteomics. How does normalisation work in Progenesis QI for proteomics? 2018. [Accessed 1 October 2018]. http://www.nonlinear.com/progenesis/qi-for-proteomics/v3.0/faq/how-normalisation-works.aspx .