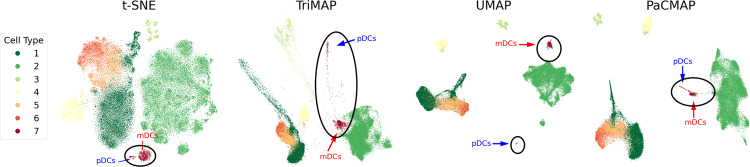
Fig. 1. Comparison of global structure preservation for DR methods.
DR visualizations of a scRNAseq dataset of PBMCs from HIV-infected people from ref. 6, pre-processed using the top 70 principal components. Labels for cell types were identified by differential expression analysis for each cluster. DC clusters are circled. Note that DR algorithms can behave differently under different random seeds, and we discuss the implication of such behavior in Supplementary Note 3 and Supplementary Fig. 5.