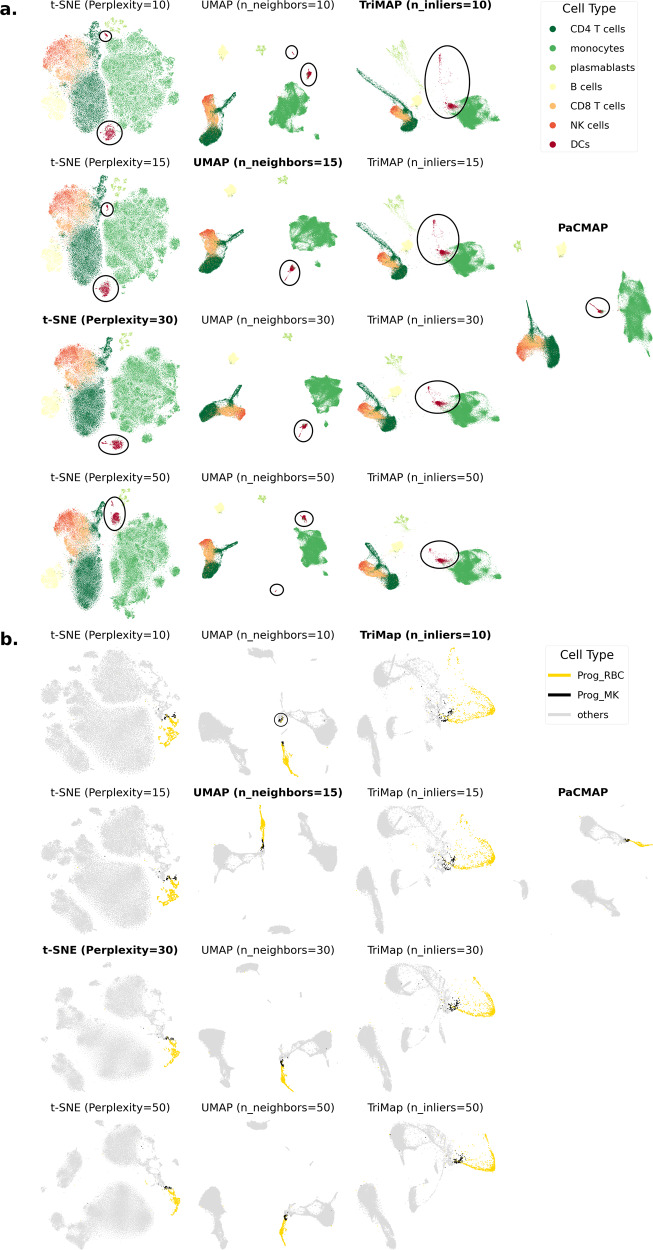
Fig. 5. Sensitivity to parameter choices on the Kazer et al.6 dataset and Stuart et al.30 dataset.
a For the Kazer et al.6 dataset, the distance relationship between DC clusters varies dramatically when tuning algorithm parameters in t-SNE and UMAP. The parameter we varied in each algorithm controls the spread of its attractive forces around each point. Clusters of DCs are circled, showing that in some cases, a single-cell type was split into two clusters by DR. The dataset was pre-processed by PCA with 70 PCs generated. b For the Stuart et al.30 dataset, the distance relationship between red blood cell progenitors (Prog_RBC) and megakaryocyte progenitors (Prog_MK) varies dramatically when tuning algorithm parameters in UMAP. Again, the parameters we varied control the spread of attractive forces. The dataset was pre-processed by PCA with 70 PCs generated.