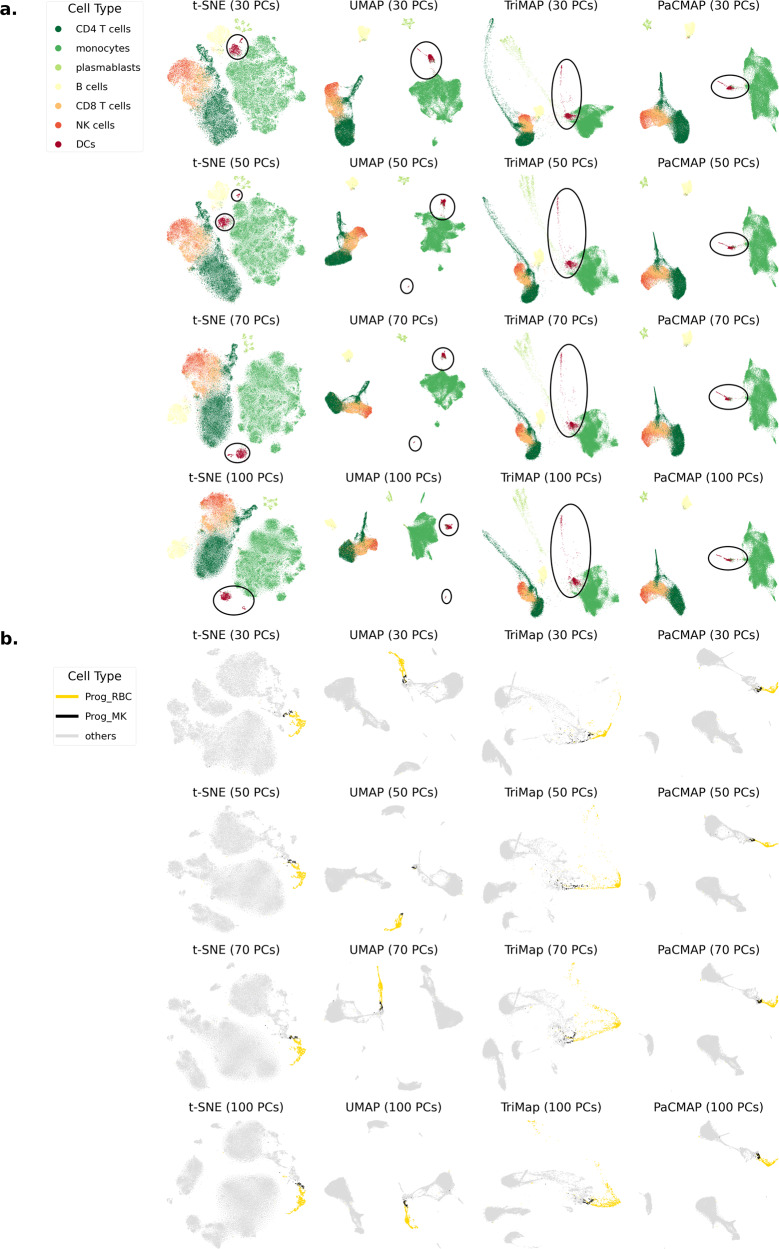
Fig. 6. Sensitivity to the number of PCA dimensions used to initialize DR.
a Here we varied the number of PCs on the Kazer et al.6 dataset. t-SNE and UMAP were not robust to the number of PCs from PCA pre-processing, having different distance relationships between two DCs clusters (pDCs and mDCs), while TriMap and PaCMAP are relatively robust in this respect. b We varied the numbers of input PCs from the Stuart et al.30 dataset. Here, UMAP is not robust to the number of PCs from PCA pre-processing, generating different results in terms of distance between red blood cell progenitors (Prog_RBC) and megakaryocyte progenitors (Prog_MK) depending on the number of PCs, while t-SNE, TriMap and PaCMAP are rather robust in this aspect. For this dataset, cell types Prog_RBC and Prog_MK should be close to each other, but UMAP with 50 PCs places points belonging to these two cell types far away from each other. Additionally, the Prog_MK cluster is separated into two clusters in UMAP's result with 50 PCs.