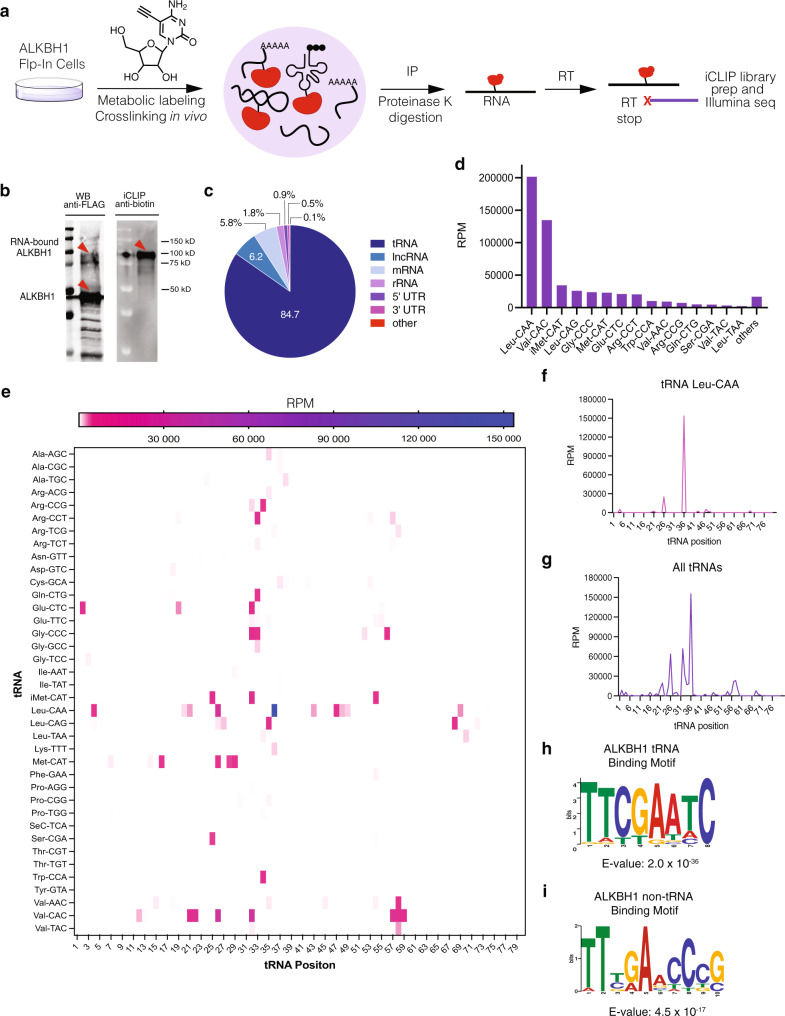
Fig. 4. 5-EC-iCLIP sequencing of ALKBH1 m5C RNA substrates.
a Strategy for 5-EC-iCLIP workflow. Cells overexpressing ALKBH1 are metabolically labeled with 5-EC and the RNA-crosslinked protein is immunoprecipitated (IP) and digested. RNA is reverse transcribed (RT) and prepared for Illumina sequencing. b Analysis of RNA-ALKBH1 crosslinking in immunoprecipitated 5-EC-iCLIP samples after biotin labeling of RNA. Experiment was repeated two times with similar results. c Distribution of uniquely mapped reads identified by 5-EC-iCLIP; lncRNA, long non-coding RNA. d Abundance of tRNA species enriched by 5-EC-iCLIP. cDNA scores from reads were normalized to reads per million (RPM). e Heatmap showing enrichment (in RPM) of all crosslink peaks in tRNA targets according to their relative position in the mature tRNAs. Peaks below 5000 RPM are not visible on color scale. f Distribution of all peaks in tRNA Leu-CAA according to their relative position in the mature tRNA. g Distribution of crosslink peaks in all tRNA targets identified by 5-EC-iCLIP according to their relative position in the mature tRNAs. h tRNA consensus motif identified by MEME from all tRNA peaks identified by 5-EC-iCLIP. i non-tRNA consensus motif identified by MEME from iCLIP non-tRNA peaks.