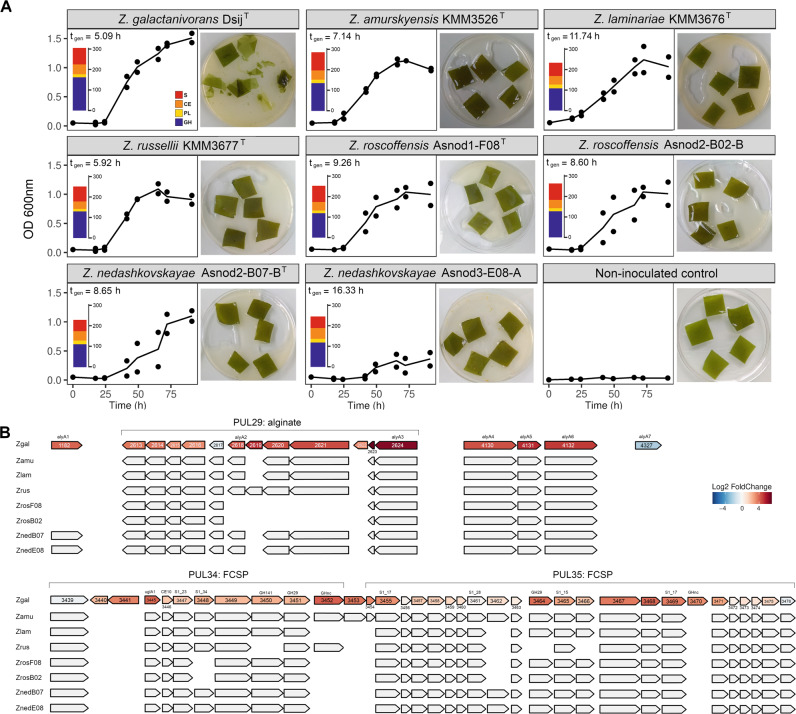
Fig. 6. All Zobellia spp. use fresh L. digitata but Z. galactanivorans DsijT displays the highest tissue breakdown capacity.
A Growth of eight Zobellia strains with L. digitata pieces (meristem of adult individuals). The generation time tgen is indicated for each strain as well as the number of glycoside hydrolases (GH, blue), polysaccharides lyases (PL, yellow), carbohydrate esterases (CE, orange) and sulfatases (S, red) predicted in their genome (dbCAN search on the MaGe platform). Individual points for duplicate experiments are shown. Lines are means of independent replicates (n = 2 or n = 3). B Comparison of genomic loci among the eight Zobellia strains. For Z. galactanivorans, genes were colored according to their expression log2FC for the comparison L. digitata vs. maltose. Gene ID is indicated inside arrows and CAZymes and sulfatases are specified above. Top: genes involved in the alginate-utilization system. Bottom: genes contained in putative FCSP PUL34 and 35. Zgal: Z. galactanivorans DsijT; Zamu: Z. amurskyensis KMM 3526 T; Zlam: Z. laminariae KMM 3676 T; Zrus: Z. russellii KMM 3677 T; ZrosF08: Z. roscoffensis Asnod1-F08T; ZrosB02: Z. roscoffensis Asnod2-B02-B; ZnedB07: Z. nedashkovskayae Asnod2-B07-BT; ZnedE08: Z. nedashkovskayae Asnod3-E08-A.