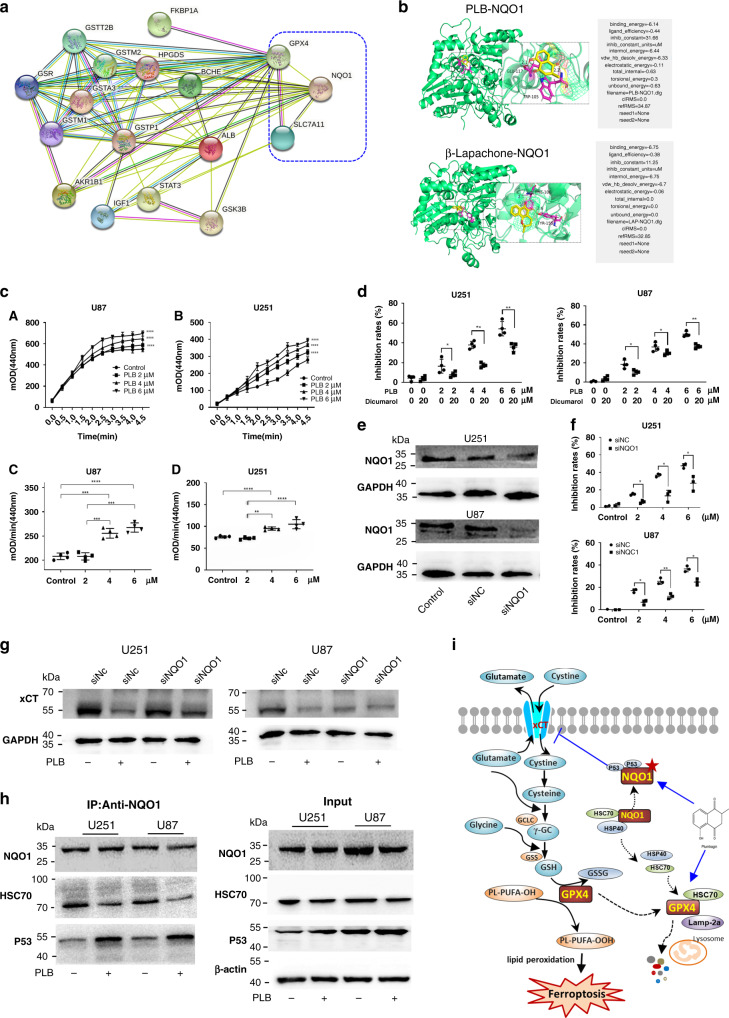
Fig. 6. PLB induced ferroptosis by targeting NQO1.
a Functional protein association network of GPX4, xCT, and PLB-related predictive targets by using the STRING database 11.0 (http://string-db.org/). PLB-related predictive targets were identified from the PharmMapper database (http://www.lilab-ecust.cn/pharmmapper/). b Molecular docking result for the binding of PLB to NQO1. β-Lapachone was used as the positive control. cA, cB The OD dynamic change curve of U251 and U87 cells with or without PLB treatment (n = 4). cC, cD Quantitation of NQO1 activity in U251 and U87 cells with or without PLB treatment (n = 4). d Cell viabilities of U251 and U87 cells treated with PLB in the presence of dicumarol (20 μM) for 48 h (n = 4). e Western blot results of NQO1 expression in siNQO1- or siNC-transfected U87 and U251 cells (n = 4). f Inhibitory effects of PLB on siNQO1- or siNC-transfected U87 and U251 cells via the MTS assay (n = 3). g Western blot results of xCT expression levels in siNQO1- or siNC-transfected U87 and U251 cells with or without 6 μM PLB treatment (n = 3). h Coimmunoprecipitation results of NQO1 and p53 or HSC70 in U251 and U87 cells treated with or without 6 μM PLB (n = 3). i Schematic of PLB-induced ferroptosis in glioma cells. Results are expressed as mean ± S.D. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.