Abstract
The immune memory to common cold coronaviruses (CCCs) influences SARS-CoV-2 infection outcome, and understanding its effect is crucial for pan-coronavirus vaccine development. We performed a longitudinal analysis of pre-COVID19-pandemic samples from 2016–2019 in young adults and assessed CCC-specific CD4+ T cell and antibody responses. Notably, CCC responses were commonly detected with comparable frequencies as with other common antigens and were sustained over time. CCC-specific CD4+ T cell responses were associated with low HLA-DR+CD38+ signals, and their magnitude did not correlate with yearly CCC infection prevalence. Similarly, CCC-specific and spike RBD-specific IgG responses were stable in time. Finally, high CCC-specific CD4+ T cell reactivity, but not antibody titers, was associated with pre-existing SARS-CoV-2 immunity. These results provide a valuable reference for understanding the immune response to endemic coronaviruses and suggest that steady and sustained CCC responses are likely from a stable pool of memory CD4+ T cells due to repeated earlier exposures and possibly occasional reinfections.
Keywords: CD4, T cells, immune memory, common cold coronaviruses, steady state, longitudinal analysis, SARS-CoV-2
Graphical abstract
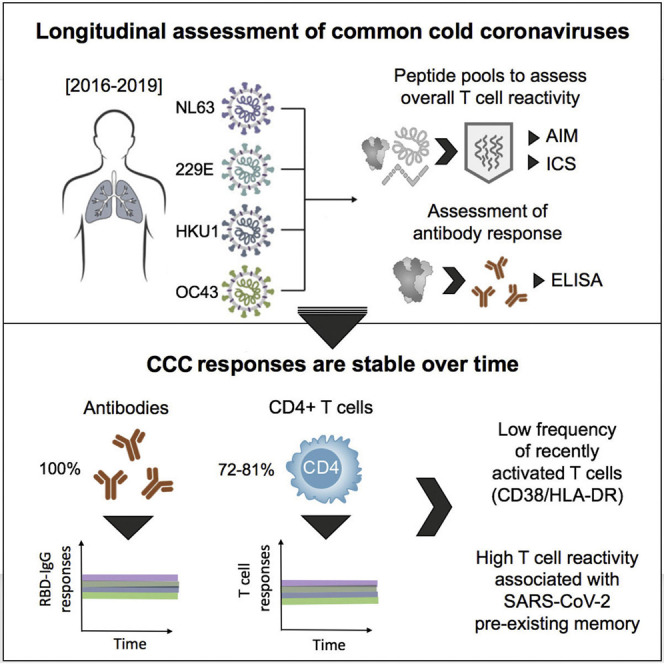
Yu et al. conducted a longitudinal study on pre-COVID pandemic samples and found that common cold coronaviruses (CCCs) responses are sustained and relatively constant over time. This “steady state” might result from both repeated infections in childhood and occasional re-exposures. It potential parallels with SARS-CoV-2 eventual transition to an endemic state.
Introduction
Common cold coronaviruses (CCCs) are seasonal viruses comprising two genera, namely, α-coronaviruses (HCoV-229E and HCoV-NL63) and β-coronaviruses (HCoV-OC43 and HCoV-HKU1), that most frequently cause mild illnesses in humans (Graat et al., 2003; Pene et al., 2003; van der Hoek et al., 2004; Walsh et al., 2013; Woo et al., 2005). CCCs are endemic viruses with widespread global distribution and have long circulated in humans. These CCC viruses are phylogenetically related to other coronaviruses that cause severe diseases in humans, such as SARS-CoV-2, SARS-CoV, and MERS-CoV (Cui et al., 2019).
CCCs have been estimated to be responsible for up to 15%–30% of pre-pandemic annual respiratory tract infections (Galanti et al., 2019; Siggins et al., 2021; Wang et al., 2020), with infections occurring most frequently in young children (Dominguez et al., 2009; Galanti et al., 2019; Selva et al., 2021). CCC infections are associated with a clear seasonality, but infection can occur at any time of the year (Killerby et al., 2018; Li et al., 2020; Park et al., 2020; Rucinski et al., 2020).
Whether immunity to CCC viruses is short or long lived has been debated with conflicting reports (Callow et al., 1990; Edridge et al., 2020s; Galanti and Shaman, 2021; Kissler et al., 2020; Petrie et al., 2021; Townsend et al., 2021; Waterlow et al., 2021). Some discrepancies may be reconciled as some reports consider immunity as protection from re-infection while others consider protection from symptomatic disease. CCC infections are associated with the generation of antibody titers widely detectable in the human population (Edridge et al., 2020; Khan et al., 2021; Lin et al., 2022; Tan et al., 2021). However, few data are available regarding the frequency of memory T cell responses against CCC and, in particular, their stability over time. Understanding the steady-state dynamics of CCC antibodies and T cell responses in humans is of potential relevance in the context of the long-term evolution of the SARS-CoV-2 during the pandemic and current scenario, where a large fraction of the human population is exposed and/or vaccinated, and several areas appear to be transitioning out of the “full-blown explosive pandemic phase” (Achenbach and Pietsch, 2022) and into a more controlled phase.
Furthermore, it has been widely reported that CCC T cell responses are associated with some degree of cross-reactivity with SARS-CoV-2 and that this cross-reactivity can at least in part explain the pre-existing T cell memory reactivity recognizing SARS-CoV-2 sequences, observed in SARS-CoV-2-unexposed subjects (Bacher et al., 2020; da Silva Antunes et al., 2021; Dykema et al., 2021; Mateus et al., 2020; Saletti et al., 2020; Tan et al., 2021; Woldemeskel et al., 2022). A putative role for CCC cross-reactive T cells in modulating COVID-19 vaccination and disease outcomes has been indicated by several independent studies (da Silva Antunes et al., 2021; Kundu et al., 2022; Loyal et al., 2021; Mateus et al., 2021; Sagar et al., 2021; Saletti et al., 2020). However, the data have not yet demonstrated which factors in a given population determine which individuals are associated with pre-existing SARS-CoV-2 T cell memory reactivity. Understanding the dynamics of CCC cross-reactivity with SARS-CoV-2 T cell responses is of potential relevance for understanding variations in COVID-19 disease severity and also relevant to the potential development of pan-coronavirus T cell vaccination (Su et al., 2022).
Herein, we performed a longitudinal analysis, over the course of 6 months up to 3 years, of CD4+ T cells and antibody responses to CCC and responses to other respiratory viruses and chronic or ubiquitous pathogens. Overall, the results suggest that responses are readily detectable and sustained over time.
Results
The frequency of CCC-specific memory CD4+ T cells is comparable to those for other common antigens
We studied peripheral blood mononuclear cells (PBMC) samples from 32 participants of a Bordetella pertussis observational study (da Silva Antunes et al., 2018). Three to seven longitudinal blood donations per donor, spanning time periods from 6 months to more than 3 years, were available. All samples were collected in the 2016–2019 period (pre-pandemic). Subjects (9 male and 23 female) represented a range of ethnicities (14 Caucasian, 10 Hispanics, 7 Asian, and 1 Black), with a median age of 24.5 years (range 18–35) (Table 1 ) and were recruited at LJI (La Jolla CA).
Table 1.
Overall characteristics of the study cohort
| Number of donors | Age (median and range) | Gender | Ethnicity | ||||
|---|---|---|---|---|---|---|---|
| – | – | Male | Female | Caucasian | Hispanic/Latino | Asian | African American |
| 32 | 24.5 (18–35) | 9 | 23 | 14 | 10 | 7 | 1 |
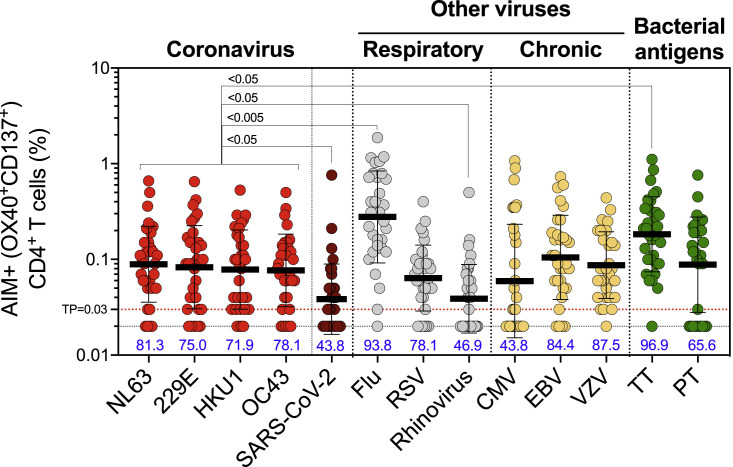
CD4+ T cell responses to the four prototypical CCC viruses (NL63, 229E, HKU1, and OC43) were measured, using the activation-induced marker (AIM) and the OX40/4-1BB markers combination (da Silva Antunes et al., 2021), which has been previously utilized to characterize viral responses and particularly SARS-CoV-2 CD4+ T cell responses (Dhanwani et al., 2021; Grifoni et al., 2020; Tarke et al., 2021; Voic et al., 2020; Yu et al., 2022). Responses to SARS-CoV-2, other respiratory viruses (influenza, respiratory syncytial virus [RSV], and rhinovirus), chronically infectious viruses (Epstein-Barr virus [EBV], cytomegalovirus [CMV], and varicella-zoster virus [VZV]), and ubiquitous bacterial vaccine antigens (Clostridium tetani [TT] and Bordetella pertussis [PT]) were measured using specific peptide sets (key resources table and method details section). CD4+ T cell responses were measured in the 32 study subjects at the first time point of the longitudinal series (Figure 1 ). Significant antigen-specific CD4+ T cell responses were detected for all four CCC epitope pools. Overall, 81.3%, 75.0%, 71.9%, and 78.1% of the donors were positive for NL63, 229E, HKU1, and OC43, respectively. The median magnitudes of the CD4+ T cell responses were 0.089%, 0.083%, 0.078%, and 0.077% for NL63, 229E, HKU1, and OC43, respectively (Figure 1). These magnitudes were 2–2.3 times significantly higher than pre-existing SARS-CoV-2 responses, which were only detected in 43.8% of the donors and consistent with previous observations (da Silva Antunes et al., 2021; Grifoni et al., 2020; Mateus et al., 2020). Similar levels of reactivity were observed when considering the stimulation index (SI) responses of ≥2 (Figure S1).
Figure 1.
CD4+ T cell responses to four representative CCCs are widely detectable in the study cohort and of similar magnitude to other pathogens
Common cold coronavirus (CCC) and several other human pathogens-specific T cell responses were measured as the percentage of AIM+ (OX40+CD137+) CD4+ T cells after stimulation of PBMCs with peptide pools. Graphs show the individual response of the four CCCs (NL63, 229E, HKU1, and OC43), SARS-CoV-2 and other pathogens plotted as background subtracted against DMSO negative control. The first time point of the longitudinal series is plotted (n = 32), and the associated percentage of positive response for each antigen is indicated. TP, threshold of positivity. Data are represented as geometric mean and SD. Kruskal-Wallis test adjusted with Dunn’s test for multiple comparisons was performed between the different antigens and each CCC virus. Adjusted p values are shown for statistically significant comparisons (p < 0.05).
The CCC-specific CD4+ T cell reactivities were in the same range as those detected for the RSV, CMV, EBV, VZV, and PT targets (Figure 1). CCC-specific CD4+ T cell reactivities were 2- to 3-fold lower than influenza (flu) (p values ranging 0.0003–0.003 and p = 0.01–0.04 for absolute and SI readouts, respectively) or TT (p values ranging 0.017–0.04 and p = 0.003–0.004 for absolute and SI readouts, respectively) responses and were 2-fold higher compared with the rhinovirus response (p values ranging 0.014–0.047 and p = 0.024–0.036 for absolute and SI readouts, respectively) (Figures 1 and S1).
The detection of CD4+ T cell responses to CCC and SARS-CoV-2 viruses was alternatively performed by intracellular cytokine staining (ICS) and the assessment of IFNγ TNFα, IL-2, and granzyme B (GzB) expression among intracellular CD154+ (CD40L) cells. EBV was used as the control. As shown in Figure S2, antigen-specific CD4+ T cell responses were readily detected by cytokine expression, and the magnitude correlated with responses measured by OX40/4-1BB markers combination. This is consistent with previous reports (Mateus et al., 2021), which also showed a high correlation between the two assays.
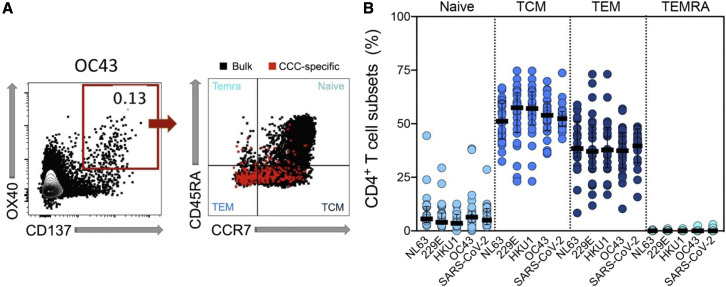
As expected, CCC- and SARS-CoV-2-specific CD4+ T cells predominantly correspond to central memory T cells (TCM) and effector memory T cells (TEM) compartments (defined as CD45RA−CCR7+ and CD45RA−CCR7−, respectively), with minimal contributions from naive (CD45RA+CCR7+) or TEMRA (CD45RA+CCR7−) compartments (Figure 2 ). Similar phenotypes were associated with the AIM+ CD4+ T cells responding to the other antigen targets (data not shown). In summary, the data demonstrate that CD4+ T cell reactivity to 229E, NL63, HKU1, and OC43 was frequently detected in the study cohort, mediated by classic conventional memory cells, and in the same order of magnitude as other viral antigens.
Figure 2.
CCC-specific CD4+ Tcells are largely classic memory cells
CCC-specific CD4+ T cell subsets (Naive: CD45RA+ CCR7+, TEMRA: CD45RA+ CCR7−, TCM: CD45RA− CCR7+, and TEM: CD45RA− CCR7−) were measured after the stimulation of PBMCs with specific peptide pools.
(A) Representative FACS plots, gated on the CCC-specific CD4+ T cells (red) measured as the percentage of AIM+ (OX40+CD137+) from total CD4 T cells (left), with the four subsets indicated in each quadrant for AIM+ cells (red) or total CD4+ T cells (black) (right) are shown.
(B) Percentages of T cell subsets from antigen-specific CD4+ T cells (OX40+CD137+) responding to the indicated pools of CCC or SARS-CoV-2, and with SI > 2 in each cohort (n = 32) at the first time point are shown. Each dot represents the response of an individual subject to an individual pool, with the median and interquartile range indicated.
Longitudinal analysis of CD4+ T cell reactivity to CCC and other antigens
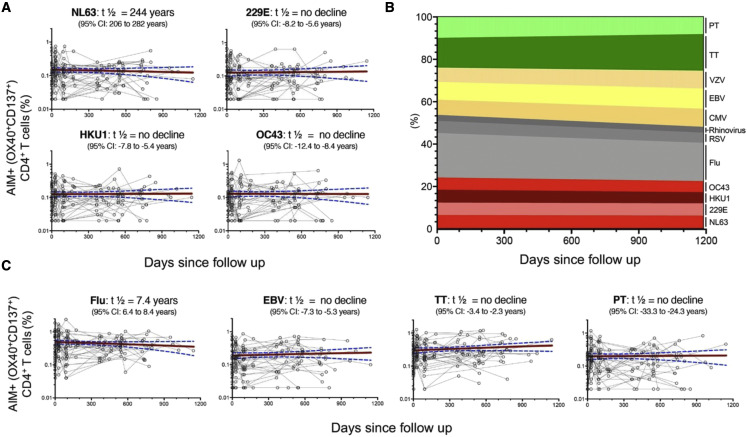
We performed a longitudinal analysis of the levels of CD4+ T cell responses to CCC and other antigens. For each antigen, the half-lives (t ½) were calculated based on linear mixed effects models using R package nlme (Cohen et al., 2021), analyzing longitudinal responses for each individual. As shown in Figure 3 A, CCC CD4+ T cell responses were essentially steady over time (t ½ ranging from 244 years to no decline). Similarly, sustained responses were observed for the other antigenic targets (Figure 3B). In particular, no decline was observed in the case of EBV, TT, and PT or a modest decline (t ½ = 7.4 years) in the case of influenza (Figure 3C). Comparable patterns were observed for RSV, rhinovirus, CMV, and VZV (Figure 3B). Overall, these results indicate relatively constant and stable responses to CCC over the time considered and are in line with what was observed for other antigens.
Figure 3.
CD4+ Tcells responses to CCC and other antigens are sustained over time
Antigen-specific T cell responses were measured as the percentage of AIM+ (OX40+CD137+) CD4+ T cells after stimulation of PBMCs with peptides pools. Individual responses of the four CCCs (A and B) or other pathogens (B and C) are shown.
(A and C) Graphs show responses plotted with all time points of the longitudinal series connected with lines for each subject (n = 32). The red line represents the median fitted curve from a nonlinear mixed effects model of longitudinal responses among those with a positive response at ≥1 time point, with 95% CI shown in blue dotted lines. t1/2 calculated based on linear mixed effects model using R package nlme (Cohen et al., 2021); t1/2 is shown as the median half-life estimated from the median slope with the associated 95% CI indicated.
(B) Longitudinal occurrence of each individual pathogen response distributed in overall percentage (sum of all absolute responses) in relation to the days since follow up.
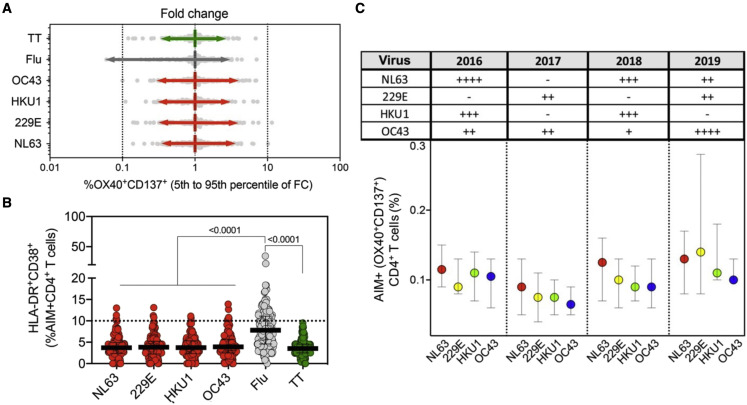
To gain more insight into whether these apparently stable responses originate from frequent reinfections or long-lasting durable responses, we determined the range of fluctuation of CD4+ T cell responses. This was done by first normalizing responses for each donor and each antigen and then calculating the associated 5th–95th percentile range. We expected that responses to influenza, where yearly vaccination/exposures are relatively common, would fluctuate more than responses to other antigens, such as TT, for which natural exposure and re-vaccination are expected to be less frequent. The data in Figure 4 A indicate that this is indeed the case. Importantly, the patterns of fluctuation of CD4+ T cell responses, more specifically, the 5th–95th percentile range for each CCC viruses (ranging 0.31–3.85) was similar to that observed for TT (0.35–2.6) and lower than what was observed in the case of influenza (0.06–2.96). These data further indicated durable and constant CD4+ T cell responses to CCC over time.
Figure 4.
CCC-specific CD4+ T cell responses are stable and not associated with recent activation or yearly changes in the prevalence of CCC infections
(A) The range of fluctuation of CD4+ T cell responses was determined by calculating the fold change of antigen-specific AIM+ (OX40+CD137+) CD4+ T cells. For each antigen, AIM+ CD4+ responses at every time point were normalized to the median of total longitudinal responses for each donor (n = 32), and the 5th–95th percentile range calculated.
(B) Graph shows CCC-, influenza-, and tetanus-specific CD4+ T cell responses associated with recent activation measured by calculating the % of HLA-DR+CD38+ from AIM+ (OX40+CD137+) CD4+ T cells at all time points of the longitudinal cohort. Each dot represents the response of an individual subject (n = 32) to an individual pool at a single time point. The median and interquartile range are represented.
(C) The prevalences of CCC infections in the West and Midwest regions during 2016–2019 were categorized according to the percent of positive rates from total tests performed (Killerby et al., 2018; Rucinski et al., 2020): −, <1%; +, 1%–2%, ++, 2%–5%; +++, 5%–8%; ++++, >8%, and results summarized in the table insert. CCC-specific CD4+ T cell responses for the four CCC were plotted as a function of the yearly incidence (2016–2019) in the graph below. Median and interquartile range are represented. Kruskal-Wallis test adjusted with Dunn’s test for multiple comparisons was performed, and the adjusted p values are shown for statistically significant comparisons (p < 0.05).
HLA-DR+CD38+ expression and periodicity of CCC antigen-specific CD4+ T cells
The expression of the HLA-DR and CD38 markers is associated with recent in vivo activation (da Silva Antunes et al., 2021; Kuri-Cervantes et al., 2020; Sekine et al., 2020). Figure 4B indicates that CD4+ T cells responding to the CCC peptides are associated with a 3.7%–3.9% range of HLA-DR+CD38+ AIM+ CD4+ T cells (95% confidence interval of 0.2%–9.6%). Only a few data points were above a threshold of high reactivity (>10%), which we previously associated with recent infection by SARS-CoV-2 of a cohort of COVID-19 convalescent subjects (da Silva Antunes et al., 2021). Interestingly, influenza-specific CD4+ T cells were associated with a median of HLA-DR+CD38+ AIM+ CD4+ T cells of 7.8% (95% confidence interval of 2.2%–16.0%), and 30% of the data points (donor/time point instances) were associated with values of 10% or higher. In the case of TT, the percent of HLA-DR+CD38+ AIM+ CD4+ T cells was similar to CCC (3.5% with a 95% confidence interval of 1.1%–7.4%), with no data point above 10%. These data are consistent with relatively more frequent exposure to influenza as compared with TT responses and are not consistent with more frequent re-exposure to CCC of the study cohort within the time frame of the longitudinal study.
The data above are derived from longitudinal samples collected in the pre-pandemic 2016–2019 calendar year period. Epidemiological data are available regarding the circulation of CCC in those years, pertaining to the West and Midwest regions (Killerby et al., 2018; Rucinski et al., 2020). If the CCC responses detected were short-lived responses resulting from frequent re-exposure, we expected that the responses would mirror the CCC circulation pattern when segregated by year. When CCC CD4+ T cell responses were plotted as a function of the year in which the blood donation was obtained, no significant associations using a multiple comparison test were observed with the yearly incidence of each individual CCC over the same period or when comparing different CCC responses within a year (Figure 4C). Overall, these data suggest that CD4+ T CCC-specific responses are not associated with recent activation or frequent yearly reinfections.
CCC circulating antibodies over time confirmed durable antibody titers and infrequent reinfections
Matched plasma samples were tested for the binding of immunoglobulin (Ig) to recombinant spike receptor binding domain (RBD) antigens from CCC as previously reported (Premkumar et al., 2020). More specifically, we measured the RBD IgG levels by the area under the curve (AUC) in titration experiments for each individual sample (Figure S3).
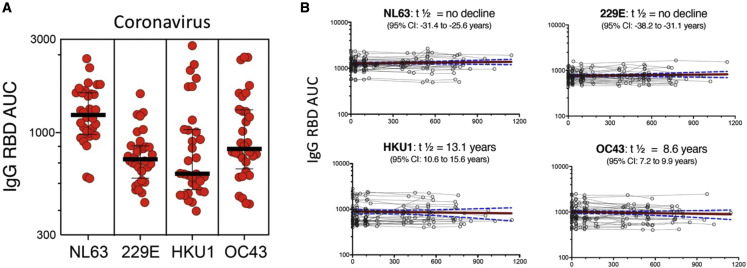
All subjects were IgG seropositive for the four CCCs (NL63, 229E, HKU1, and OC43) at the first and subsequent blood donations, consistent with their high prevalence in human adult populations (Edridge et al., 2020; Killerby et al., 2018; Nickbakhsh et al., 2020) (Figure 5 A). Durability assessments of circulating antibody titers were performed as based curve fits to model (Cohen et al., 2021), similarly to the assessments of CD4+ T cells responses above. CCC titers were sustained over time for all the 4 CCCs (Figure 5B). NL63 and 229E titers showed no decline throughout the study, whereas a modest decline was observed for HKU1 and OC43 with a t1/2 of 13.1 and 8.6 years, respectively. The stability of CCC-specific IgG responses indicates that the cohort analyzed was not associated with frequent reinfections during the time considered in the study.
Figure 5.
CCC-specific IgG responses are detected in all individuals and sustained over time
(A) Plasma IgG titers, measured by the AUC, to the spike receptor binding domain (RBD) protein of the CCC viruses (HcoV-229E, HcoV-NL63, HcoV-HKU1, and HcoV-OC43) are shown for first time point of the longitudinal cohort (n = 32). Geometric mean titers with SD are indicated.
(B) Graphs show individual CCC antibody responses plotted for all time points of the longitudinal series and connected with lines for each subject (n = 32). The red line represents the median fitted curve from a nonlinear mixed effects model of longitudinal responses among those with a positive response at ≥1 time point, with 95% CI shown in blue dotted lines. t1/2 calculated based on linear mixed effects model using R package nlme (Cohen et al., 2021); t1/2 is shown as the median half-life estimated from the median slope with the associated 95% CI indicated.
Correlation of CCC-specific CD4+ T cell and SARS-CoV-2 pre-existing responses
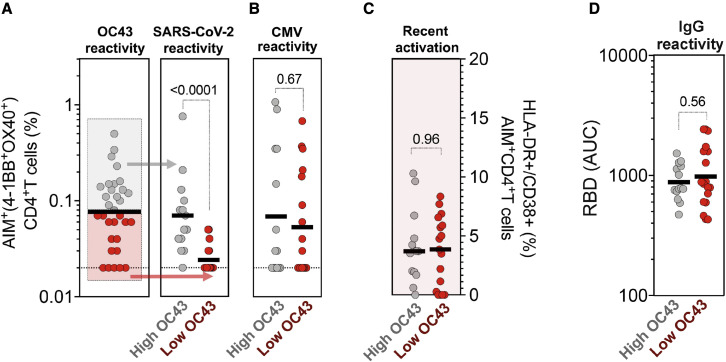
CCCs have significant sequence homology to SARS-CoV-2. The cross-reactive CCC-specific T cell responses with SARS-CoV-2 (da Silva Antunes et al., 2021; Low et al., 2021; Mateus et al., 2020) may contribute to the modulation of SARS-CoV-2 infection and enhance responses to COVID-19 vaccination (da Silva Antunes et al., 2021; Kundu et al., 2022; Loyal et al., 2021; Mateus et al., 2021; Sagar et al., 2021). We show that high CD4+ T cell memory OC43 reactivity is associated with higher levels of pre-existing memory reactivity to SARS-CoV-2 (Figure 6 A). Similar patterns were observed for NL63, 229E, and HKU1 (Figure S4) but not for the unrelated and ubiquitous pathogen CMV (Figure 6B). Consistent with this notion, the magnitude of CD4+ T cell responses specific for each individual CCC species detected in the study in the various individual subjects and time points correlated with SARS-CoV-2 responses and with each other, but not CMV (Figure S5), which is consistent (but does not prove) cross-reactivity. The highest levels of pre-existing SARS-CoV-2 reactivity were not associated with higher levels of HLA-DR/CD38+ expression (Figure 6C) suggesting that SARS-CoV-2 cross-reactive cells are not the result of a recent activation or infection. Similarly, high pre-existing SARS-CoV-2 CD4+T cell responses did not correlate with higher IgG antibody reactivity (Figure 6D). Overall, these results suggest that high CD4+ T cell reactivity to CCC, but not antibody reactivity, is predictive of cross-reactive SARS-CoV-2 CD4+ T cell responses.
Figure 6.
High CD4+ T cell reactivity to OC43 is associated with high pre-existing SARS-CoV-2 immunity
(A and B) Antigen-specific T cell responses were measured as the percentage of AIM+ (OX40+CD137+) CD4+ T cells after stimulation of PBMCs with peptides pools for (A) CCC (OC43) and SARS-CoV-2 (representing pre-existing immunity in pre-pandemic samples), (B) CMV as a control.
(C) Recent activated CCC (OC43) specific T cell responses were measured by calculating the percent of HLA-DR+CD38+ of AIM+ (OX40+CD137+) CD4+ T cells.
(D) Plasma IgG titers to CCC viruses (OC43) spike receptor binding domain (RBD) protein were measured by ELISA.
(A–D) Each dot represents the response of an individual subject (n = 32) at the first time point with the median bar shown. High responders for OC43 (above the median bar in A) are shown in gray, and low responders for OC43 (below the median bar in A) are shown in red. The different immune responses between high and low responders were compared using Mann-Whitney test, and p values < 0.05 are considered statistically significant.
Discussion
Despite the high prevalence of CCC infection, most adults experience asymptomatic or mild common cold symptoms. Although seroconversion to CCC is near ubiquitous during childhood, little is known about the dynamics of CCC-specific memory responses in adults.
In this study, our goal was to characterize CD4+ T cell-specific memory responses to the four prototypic endemic and widely circulating CCC viruses in a longitudinal cohort, using a strategy based on ex vivo stimulation of PBMCs with peptide pools covering the entire proteome of each individual virus. In particular, our data show that T cell reactivity against CCC is detected for most subjects and that this reactivity is similar in magnitude to other antigens, sustained and relatively constant over time. These observations were paralleled by equally stable circulating spike RBD antibody titers. Although we cannot exclude that at least some reinfections might have occurred during the longitudinal follow-up period, the preponderance of evidence suggests that the reactivity observed is associated with memory and persistent responses.
Previous work has suggested that substantial immune memory involving both the arms of adaptive immune memory is generated against CCC (da Silva Antunes et al., 2021; Edridge et al., 2020; Khan et al., 2021; Tan et al., 2021). We observed that about 72%–81% of subjects exhibited modest immune CD4+ T cell memory responses to each of the 4 CCC studied, with frequencies and magnitudes consistent with values detected in both community and health care workers cohorts (da Silva Antunes et al., 2021), similar to the findings of Saletti et al. (2020) and Tan et al. (2021). Saletti et al. (2020) used ELISpot to define CCC-specific responses and showed that these were weaker in older donors. There was some modest degree of cross-reactivity with SARS-CoV-2. The magnitude of CCC-specific responses is also comparable with the ubiquitous and immunodominant pathogen CMV. More specifically, using similar methodologies, other studies showed comparable levels of CMV-specific CD4+ T cell reactivity in the general population (da Silva Antunes et al., 2021; Mateus et al., 2021), and the study of Dhanwani et al. (2021) showed a similar range of CMV-specific responses in CMV+ individuals.
Remarkably, we found a stable and sustained T cell and antibody responses against CCC that are supported by recent experimental findings from Cohen et al. (2021) or mathematical modeling (Townsend et al., 2021; Waterlow et al., 2021) and argues against the short-lived nature of CCC responses (Callow et al., 1990; Edridge et al., 2020; Kissler et al., 2020).
Our findings are also in agreement with the stability of responses against other viral infections, such as vaccinia or SARS-CoV-1 where antigen-specific cells were detectable 50 years and 17 years post infection, respectively (Demkowicz et al., 1996; Le Bert et al., 2020), and against TT which are remarkably stable for many years upon vaccination (Hammarlund et al., 2016). It is quite possible that the CCC “steady-state” reactivity might be a result of both repeated infections in early childhood and occasional re-exposure and re-infection. People are known to be reinfected by CCC, and these reinfections resulted in the boosting of the CCC-specific antibody responses (Edridge et al., 2020). More specifically, as shown in Table 1 of the paper, the study relates to a total of 205.6 years of follow up, and during that period, a total of 25 NL63 infections were detected, resulting in a frequency of infection of one new infection every 8.2 years (205.6/25 = 8.2). By similar calculations based on the data shown in Table 1 of the Edridge et al. (2020) paper, the frequency of new infections for 229E, OC43, and HKU1 are one new infection every 5.4, 6.9, and 25.7 years, respectively. In our study, we examined longitudinal responses within a 3-year period, and it is therefore not unexpected that reinfections would not be highly prevalent during the follow-up period.
The data presented herein are also relevant in the context of CCC cross-reactivity with SARS-CoV-2. It has been shown that pre-existing T cell immunity elicited by past CCC exposures can influence COVID-19 responsiveness to vaccination and disease outcome (da Silva Antunes et al., 2021; Kundu et al., 2022; Loyal et al., 2021; Mateus et al., 2021; Sagar et al., 2021 and 2020). We found that although everybody had detectable antibody titers to CCC, individuals varied in the level of T cell reactivity and that the subjects with high CCC T cell reactivity, but not antibody titers, are those most likely to be associated with pre-existing SARS-CoV-2 immune reactivity. This is consistent with findings that CCC antibodies might not protect against SARS-CoV-2 infection or disease severity (Lin et al., 2022; Wratil et al., 2021), but T cells do (Kundu et al., 2022; Mallajosyula et al., 2021). These data also suggest that the degree of sequence homology between CCC and SARS-CoV-2 account for the pre-existing T cell memory reactivity of SARS-CoV-2 in some, but not all, individuals and that other factors such as recent exposure, HLA type, or other individual and environmental factors might play a role. The paper by Bacher et al. (2020) also looked at CCC-specific CD4+ responses and identified memory responses, but these were poorly cross-reactive against SARS-CoV-2.
SARS-CoV-2 antibody and T cell effector activity contract over time, and protection from infection wanes, but protection from severe disease appears to be preserved to a significant degree (Dan et al., 2021; Ortega et al., 2021; Sette and Crotty, 2021; Siggins et al., 2021). Likewise, it has also been suggested that recurrent CCC infections are only rarely associated with moderate or severe clinical symptoms (Edridge et al., 2020; Galanti and Shaman, 2021). In the context of the recent debate on the evolution of SARS-CoV-2 during the COVID19 pandemic, our data provide a glimpse of how in the young adult population, a sizeable and durable immunity to coronaviruses is sustained over years, presumably as a result of repeated earlier exposures, and possibly occasional reinfections. The development of pan-coronavirus vaccines that target not only SARS-CoV-2 but also CCC viruses might contribute to further protection.
In summary, we found that in addition to widespread antibody reactivity to all the four CCCs, memory T cell responses are detected for most individuals, and their reactivity remained stable and relatively constant over time. The characterization of the immune response to the prevalent and endemic CCCs provides a valuable reference for understanding the durability and eventual transition to an endemic state of SARS-CoV-2 in the aftermath of the pandemic.
Limitations of the study
A limitation of this investigation is the unknown history of previous CCC exposure of the study participants. The assessment of CCC infection by RT-PCR was not part of the original study design for the study and would also not have been logistically feasible over the longitudinal course of the study as it would have required frequent nasal swabs of all subjects. Therefore, in this study, the stability of T cell and antibody responses could not be directly correlated with protection from symptomatic colds and/or infection. Furthermore, our analysis is limited to “steady-state” responses in adults in a relatively short follow-up time, and the evolution of CCC responses in children until adulthood was not addressed. Although these findings were validated with several different approaches, the work is mostly limited to AIM-assessed peptide responses, and further assessment against specific epitopes and viral targets or in additional assays, such as ELISpot, would be helpful to support evidence for cross-reactivity, as performed in other studies (Mateus et al., 2020; Saletti et al., 2020). Additional limitations of this study are the relatively small size and narrow age of the cohort investigated that included only young adults and no adults of middle or advanced age. Both the frequency of CCC infections and immune memory responses to CCC may differ in older age groups. The validation of the results in age and geographically distinct populations would be desirable to generalize the findings broadly. Serology for other pathogens besides CCC was also not performed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies for AIM T cell assay | ||
| anti-CD3 (BV805) (UCHT1) | BD Biosciences | Cat#: 612895; RRID: |
| anti-CD4 (BV605) (RPA-T4) | BD Biosciences | Cat#: 562658; RRID: AB_2737935 |
| anti-CD8 (BUV496) (RPA-T8) | BD Biosciences | Cat#: 612942; RRID: AB_2563505 |
| anti-CD14 (V500) (M5E2) | BD Biosciences | Cat#: 561391; RRID: AB_10611856 |
| anti-CD19 (V500) (HIB19) | BD Biosciences | Cat#: 561121; RRID: AB_10562391 |
| anti-CD137 (APC) (4B4-1) | Biolegend | Cat#: 309810; RRID: AB_830672 |
| anti-CD134 (PE-Cy7) (Ber-ACT35) | Biolegend | Cat#: 350012; RRID: AB_10901161 |
| anti-CD69 (PE) (FN50) | BD Biosciences | Cat#: 555531; RRID: AB_2737680 |
| anti-CD45RA (BV421) (HI100) | Biolegend | Cat#: 304130; RRID: AB_10965547 |
| anti-CCR7 (FITC) (G043H7) | Biolegend | Cat#: 353216; RRID: AB_10916386 |
| Live/Dead Viability (eF506/Aqua) | Invitrogen | Cat#: 65-0866-18; RRID: N/A |
| Antibodies for ICS T cell assay | ||
| anti-CD3 (BV805) (UCHT1) | BD Biosciences | Cat#: 612895; RRID: AB_2870183 |
| anti-CD4 (BV605) (RPA-T4) | BD Biosciences | Cat#: 562658; RRID: AB_2737935 |
| anti-CD8 (BUV496) (RPA-T8) | BD Biosciences | Cat#: 612942; RRID: AB_2563505 |
| anti-CD14 (V500) (M5E2) | BD Biosciences | Cat#: 561391; RRID: AB_10611856 |
| anti-CD19 (V500) (HIB19) | BD Biosciences | Cat#: 561121; RRID: AB_10562391 |
| anti-CD137 (APC) (4B4-1) | Biolegend | Cat#: 309810; RRID: AB_830672 |
| anti-CD134 (PE-Cy7) (Ber-ACT35) | Biolegend | Cat#: 350012; RRID: AB_10901161 |
| anti-CD69 (PE) (FN50) | BD Biosciences | Cat#: 555531; RRID: AB_2737680 |
| Live/Dead Viability (eF506/Aqua) | Invitrogen | Cat#: 65-0866-18; RRID: N/A |
| anti-IFNγ (FITC) (4S.B3) | Invitrogen | Cat#: 11-7319-82; RRID: AB_465415 |
| anti-TNFα (eFluor450) (MAb11) | Life Tech | Cat#: 48-7349-42; RRID: AB_2043889 |
| anti-IL-2 (BB700) (MQ1-17H12) | BD Biosciences | Cat#: 566405; RRID: AB_2744488 |
| anti-Granzyme B (AF700) (GB11) | BD Biosciences | Cat#: 560213; RRID: AB_1645453 |
| anti-CD154 (APC-ef780) (24-31) | eBioscience | Cat#: 47-1548-42; RRID: AB_1603203 |
| Biological samples | ||
| Human blood samples | La Jolla Institute for Immunology | https://www.lji.org |
| Chemicals, peptides, and recombinant proteins | ||
| HCoV-NL63 peptides: NL63 (280 peptides) | (da Silva Antunes et al., 2021) | N/A |
| HCoV-229E peptides: 229E (225 peptides) | (da Silva Antunes et al., 2021) | N/A |
| HCoV-HKU1 peptides: HKU1 (320 peptides) | (da Silva Antunes et al., 2021) | N/A |
| HCoV-OC43 peptides: OC43 294 peptides) | (da Silva Antunes et al., 2021) | N/A |
| SARS-CoV-2 peptides: SARS-CoV-2 (474 peptides) | (da Silva Antunes et al., 2021) | N/A |
| Cytomegalovirus peptides: CMV (313 peptides) | (Dhanda et al., 2019) | N/A |
| Epstein-Barr virus peptides: EBV (301 peptides) | (Dhanda et al., 2019) | N/A |
| Influenza A peptides: Flu (330 peptides) | (Dhanda et al., 2019) | N/A |
| Respiratory syncytial virus peptides: RSV (216 peptides) | (Dhanda et al., 2019) | N/A |
| Rhinovirus peptides: Rhinovirus (136 peptides) | (Grifoni et al., 2019) | N/A |
| Varicella zoster virus peptides: VZV (335 peptides) | (Voic et al., 2020) | N/A |
| Clostridium tetani peptides: TT (125 peptides) | (da Silva Antunes et al., 2017) | N/A |
| Bordetella pertussis peptides: PT (132 peptides) | (da Silva Antunes et al., 2020) | N/A |
| Software and algorithms | ||
| GraphPad Prism Version 9 | GraphPad Software | https://www.graphpad.com |
| Microsoft Excel Version 16.16.27 | Microsoft | https://www.microsoft.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact: Dr. Ricardo da Silva Antunes (rantunes@lji.org).
Materials availability
Epitope pools used in this study will be made available to the scientific community upon request, and following execution of a material transfer agreement (MTA), by contacting A.S. (alex@lji.org) and R.d.S.A (rantunes@lji.org). Likewise, biomaterials archived from this study may be shared for further research with MTA.
Experimental model and subject details
Study cohort and PBMC isolation
The purpose of this study was to investigate the immunological memory to common cold corona viruses in a longitudinal cohort collected during pre-pandemic time. Blood donations from 32 donors previously recruited in a Bordetella Pertussis observational study were collected under IRB approved protocols at the La Jolla Institute for Immunology (protocol no. VD-101), before COVID-19 pandemic from 2016-2019. There were 3 to 7 longitudinal blood donations per donor spanning time periods from 6 months to more than 3 years. Each participant provided informed consent and was assigned a study identification number with clinical information recorded. Exclusion criteria included pregnancy at the start of the study; presentation of severe disease; medical treatment that might interfere with study results and/or antibiotic use or fever (>100.4°F [38°C]). Adults of all races, ethnicities, ages, and genders were eligible to participate, and the aggregate information can be found in Table 1. The association of gender on the results of the study was not explicitly measured. In all cases, PBMCs were isolated from whole blood by density gradient centrifugation according to manufacturer instructions (Ficoll-Hypaque, Amersham Biosciences, Uppsala, Sweden) and cryopreserved for further analysis.
Study approval
This study was approved under IRB protocol approval (VD-101) at the La Jolla Institute for Immunology. All donors were able to provide informed consent, or had a legal guardian or representative able to do so. Each participant provided informed consent and was assigned a study identification number with clinical information recorded.
Method details
Synthesis of epitope pools
In the current study, in order to investigate the CCC and SARS-CoV-2 specific T cell responses, we used megapools (MPs) combing the overlapping spike (S) epitope pools and predicted HLA class II CD4+ T cell epitope pools from the rest of the genome (R) (key resources table), generated using previously described strategies (da Silva Antunes et al., 2021; Grifoni et al., 2020) utilizing the Immune Epitope Database and Analysis Resource (IEDB). Briefly, we generated MPs of 15-mer peptides overlapping by 10 spanning the entire Spike protein sequences or alternatively MPs for the remainder genomes consisting of dominant HLA class II predicted CD4+ T-cell epitopes. We also studied antigen-specific responses against a panel of other respiratory viruses (influenza, RSV, and rhinovirus), chronically infectious viruses (EBV, CMV, and VZV), and ubiquitous bacterial vaccine antigens (TT and PT) using peptide sets described in key resources table. Detailed overall information of the MPs composition, peptide numbers as well as references are specified in key resources table. Individual peptides were synthesized by TC peptide lab (San Diego, CA) and pooled by protein combinations and resuspended to a final concentration of 1 mg/mL in DMSO.
CCC Spike protein RBD enzyme-linked immunosorbent assay
All plasma samples tested by ELISA assay were heat-inactivated at 56°C for 30 min to reduce risk from any possible residual virus in serum. Briefly, 50 μL of Streptavidin (Invitrogen) at 4 μg/mL in Tris-Buffered Saline (TBS) pH 7.4 was coated in the 96-well, high-binding microtiter assay plate (Greiner Bio-One cat # 655061) for 1 hour at 37°C. The coating solution was removed, then 100 μL of blocking solution, 1:1 Non Animal Protein-BLOCKER™ (G-Biosciences) in TBS was added for 1 hour at 37°C. Serum samples were serially diluted (1:40 – 1:8100), in 3% Bovine Serum Albumin (BSA) in TBS containing 0.05% Tween 20 (TBST) with respective biotinylated spike RBD antigens from CCC at 1 μg/mL in a 96-round-well V bottom plate (Diaago cat # R96-300V) and incubated for 1 hour at 37°C. The blocking solution was removed, then 50 μL of diluted serum was added to the assay plate and incubated for 15 minutes at 37°C. The plate was washed three times using wash buffer (1X TBS containing 0.2% Tween 20), then 50 μL of horseradish peroxidase-conjugated secondary Goat Anti-Human secondary IgG antibody (Cat No: 109-035-008, Jackson ImmunoResearch) at 1:40,000 dilution in 3% milk was added for 1 hour at 37°C. The plate was washed three times using wash buffer, then 50 μL of 3,3’,5,5’ -Tetramethylbenzidine (TMB) Liquid Substrate (Sigma-Aldrich) was added to the plate, and absorbance was measured at 450 nm using a plate reader (Molecular Devices SpectraMax ABS Plus Absorbance ELISA Mcroplate Reader) after stopping the reaction with 50 μl of 1 N HCl. Area under the curve for titration experiments for each sample were calculated by the trapezoidal model implemented in Prism Version 9.3.0.
Activation induced cell marker (AIM) T cell assay
The AIM assay was performed as previously described (Mateus et al., 2020). Cryopreserved PBMCs were thawed by diluting the cells in 10 mL complete RPMI 1640 with 5% human AB serum (Gemini Bioproducts) in the presence of benzonase [20 ml/10ml]. Cells were cultured for 20 to 24 hours in the presence of CCC or SARS-CoV-2 specific and other common antigen pools (1ug/ml) in 96-wells U bottom plates with 1x106 PBMC per well. An equimolar amount of DMSO was added as a negative control and phytohemagglutinin (PHA, Roche (San Diego, CA) 1 mg/ml) was used as the positive control. Cells were stained and activation of CD4+ T cell measured by the CD137 and OX40 marker combination. The detailed information of all the antibodies used are summarized in key resources table. All samples were acquired on a ZE5 cell analyzer (Biorad laboratories, Hercules, CA) and analyzed with FlowJo software (Tree Star, Ashland, OR). AIM+ CD4+ T cells data were calculated as percent of total CD4+ T cells background subtracted or stimulation index. Background subtracted data were derived by subtracting the percentage of AIM+ cells percentage after each MP stimulation from the DMSO stimulation. The Stimulation Index (SI) was calculated by dividing the count of AIM+ cells after SARS-CoV-2 pools stimulation with the ones in the negative control. A positive response was defined as SI greater than 2 and AIM+ response above the threshold of positivity after background subtraction. The limit of detection (0.02%) was calculated based on 2 times 95% CI of geomean of negative control (DMSO), and the threshold of positivity (0.03%) was calculated based on 2 times standard deviation of background signals according to previous published studies (Dan et al., 2021; Mateus et al., 2020). All data below 0.02 or SI<2 were set to 0.02 or 2 for plotting and statistical analysis. The detailed gating strategy used to define CD4+ AIM reactive cells (OX40+CD137+), memory (CD45RA/CCR7), and activated sub-populations (HLA-DR+CD38+) is listed in Figure S6. Gates were drawn relative to the unstimulated condition for each donor.
Intra cellular staining (ICS) T cell assay
The ICS T cell assay was performed as previously described (Tarke et al., 2022). PBMCs were cultured in the presence of antigen-specific MPs [1 mg/ml] in 96-well U-bottom plates at a concentration of 2x106 PBMC per well. As a negative control, an equimolar amount of DMSO was used to stimulate the cells in triplicate wells and phytohemagglutinin (PHA, Roche, (San Diego, CA) 1mg/ml) stimulated cells were used as positive controls. After incubation for 24 hours at 37oC in 5% CO2, cells were incubated for additional 4 hours after adding Golgi-Plug containing brefeldin A, Golgi-Stop containing monensin (BD Biosciences, San Diego, CA) together with CD137 APC antibody (2:100; Biolegend, San Diego, CA). Cells were then stained on their surface for 30 min at 4oC in the dark, after that fixed with 1% of paraformaldehyde (Sigma-Aldrich, St. Louis, MO), permeabilized, and blocked for 15 minutes followed by intracellular staining for 30 min at room temperature. The detailed information of the antibodies used in ICS assay are summarized in key resources table. All samples were acquired on a ZE5 5-laser cell analyzer (Biorad laboratories, Hercules, CA) and analyzed with FlowJo software (Tree Star, Ashland, OR). The gates for cytokine positive cells were drawn relative to the negative and positive controls for each donor. Specifically, lymphocytes were gated, followed by single cells determination. T cells were gated for being positive to CD3 and negative for a Dump channel including in the same colors CD14, CD19 and Live/Dead staining. CD3+CD4+ and CD3+CD8+ cells were further gated based on a combination of each cytokine (IFNg, TNFa, IL-2, Granzyme B) with CD40L (CD154). The total cytokine response and T cell functionality was calculated from Boolean gating of single cytokines that was applied to CD3+CD4+ cells. The background was removed from the data by subtracting the average of the % of Cytokine+ cells plated in triplicate wells stimulated with DMSO. For ICS, CD4+ T cell responses were based on the expression of CD40L (CD154) in combination with IFNg, TNFa, IL-2 or Granzyme B, the sum of the double positive represents the overall CD4+Cytokine+. CD4 cytokine responses were background subtracted individually and found positive only if fulfilling the criteria of an SI greater than 2 and a threshold of positivity (TP) of 0.005% single cytokine CD4+ T cells (0.02% for overall CD4+Cytokine+ cells). The TP for ICS was considered to be a positive response based on the median twofold standard deviation of T cell reactivity in negative DMSO controls. The detailed gating strategy and cytokine detection for a representative donor is shown in Figure S6. Gates were drawn relative to the unstimulated condition for each donor.
Quantification and statistical analysis
Experimental data were analyzed by GraphPad Prism Version 9 (La Jolla, CA) and Microsoft Excel Version 16.16.27 (Microsoft, Redmond, WA). The statistical details of the experiments are provided in the respective figure legends. Data were analyzed by Mann-Whitney test (two-tailed) to compare between two groups, and Kruskal-Wallis test adjusted with Dunn’s test for multiple comparisons to compare between multiple groups. The regression lines and estimated t ½ was calculated based on linear mixed effects model using R package nlme as previously described by Cohen et al. (2021). Data were plotted as geometric mean with geometric SD for log scale and median with interquartile range for numeric scale. p values < 0.05 (after adjustment if indicated) were considered statistically significant.
Acknowledgments
We wish to acknowledge all the subjects for their participation and for donating their blood and time for this study. We are grateful to the La Jolla Institute for Immunology Clinical core’s relentless efforts in obtaining blood samples. The research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under award numbers U01AI141995, U19AI142742, and U54CA260543 and contract number 75N93019C00066. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Designing research studies, A.S., R.d.S.A., and E.D.Y.; investigation, A.S., R.d.S.A., E.D.Y., T.M.N., E.W., E.G., L.P., and J.M.; data analysis, E.D.Y., R.d.S.A., and L.P.; resources, A.S. and R.d.S.A.; manuscript writing, A.S., R.d.S.A., E.D.Y., L.P., D.W., and A.G.; supervision, A.S. and R.d.S.A.; project administration, A.F.; funding acquisition, A.S. and R.d.S.A.
Declaration of interests
A.S. is a consultant for Gritstone Bio, Flow Pharma, Arcturus Therapeutics, ImmunoScape, CellCarta, Avalia, Moderna, Fortress, and Repertoire. L.J.I. has filed for patent protection for various aspects of the SARS-CoV-2 epitope pools design.
Published: July 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.07.012.
Supplemental information
Data and code availability
-
•
This paper uses publicly available datasets for epitope synthesis. Their references are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/k76f5sjjmh.1 Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request
References
- Bacher P., Rosati E., Esser D., Martini G.R., Saggau C., Schiminsky E., Dargvainiene J., Schröder I., Wieters I., Khodamoradi Y., et al. Low-avidity CD4+ T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. 2020;53:1258–1271.e5. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen K.W., Linderman S.L., Moodie Z., Czartoski J., Lai L., Mantus G., Norwood C., Nyhoff L.E., Edara V.V., Floyd K., et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell. Rep. Med. 2021;2:100354. doi: 10.1016/j.xcrm.2021.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R., Babor M., Carpenter C., Khalil N., Cortese M., Mentzer A.J., Seumois G., Petro C.D., Purcell L.A., Vijayanand P., et al. Th1/Th17 polarization persists following whole-cell pertussis vaccination despite repeated acellular boosters. J. Clin. Invest. 2018;128:3853–3865. doi: 10.1172/JCI121309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R., Pallikkuth S., Williams E., Dawen Yu E., Mateus J., Quiambao L., Wang E., Rawlings S.A., Stadlbauer D., Jiang K., et al. Differential T-cell reactivity to endemic coronaviruses and SARS-CoV-2 in community and health care workers. J. Infect. Dis. 2021;224:70–80. doi: 10.1093/infdis/jiab176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R., Paul S., Sidney J., Weiskopf D., Dan J.M., Phillips E., Mallal S., Crotty S., Sette A., Lindestam Arlehamn C.S. Definition of human epitopes recognized in tetanus toxoid and development of an assay strategy to detect ex vivo tetanus CD4+ T cell responses. PLoS One. 2017;12:e0169086. doi: 10.1371/journal.pone.0169086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Antunes R., Quiambao L.G., Sutherland A., Soldevila F., Dhanda S.K., Armstrong S.K., Brickman T.J., Merkel T., Peters B., Sette A. Development and validation of a Bordetella pertussis whole-genome screening strategy. J. Immunol. Res. 2020;2020:8202067. doi: 10.1155/2020/8202067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demkowicz W.E., Jr., Littaua R.A., Wang J., Ennis F.A. Human cytotoxic T-cell memory: long-lived responses to vaccinia virus. J. Virol. 1996;70:2627–2631. doi: 10.1128/jvi.70.4.2627-2631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda S.K., Mahajan S., Paul S., Yan Z., Kim H., Jespersen M.C., Jurtz V., Andreatta M., Greenbaum J.A., Marcatili P., et al. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019;47:W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanwani R., Dhanda S.K., Pham J., Williams G.P., Sidney J., Grifoni A., Picarda G., Lindestam Arlehamn C.S., Sette A., Benedict C.A. Profiling human Cytomegalovirus-specific T cell responses reveals novel immunogenic open reading frames. J. Virol. 2021;95:e0094021. doi: 10.1128/JVI.00940-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez S.R., Robinson C.C., Holmes K.V. Detection of four human coronaviruses in respiratory infections in children: a one-year study in Colorado. J. Med. Virol. 2009;81:1597–1604. doi: 10.1002/jmv.21541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykema A.G., Zhang B., Woldemeskel B.A., Garliss C.C., Cheung L.S., Choudhury D., Zhang J., Aparicio L., Bom S., Rashid R., et al. Functional characterization of CD4+ T cell receptors crossreactive for SARS-CoV-2 and endemic coronaviruses. J. Clin. Invest. 2021;131:e146922. doi: 10.1172/JCI146922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Achenbach J., Pietsch B. U.S. no longer in ‘full-blown’ pandemic phase, Fauci says. Washington Post. 2022. https://www.washingtonpost.com/health/2022/04/27/pandemic-phase-over-fauci-covid/ April 27, 2022.
- Galanti M., Birger R., Ud-Dean M., Filip I., Morita H., Comito D., Anthony S., Freyer G.A., Ibrahim S., Lane B., et al. Longitudinal active sampling for respiratory viral infections across age groups. Influenza Other Respir. Viruses. 2019;13:226–232. doi: 10.1111/irv.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanti M., Shaman J. Direct observation of repeated infections With endemic coronaviruses. J. Infect. Dis. 2021;223:409–415. doi: 10.1093/infdis/jiaa392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graat J.M., Schouten E.G., Heijnen M.L., Kok F.J., Pallast E.G., de Greeff S.C., Dorigo-Zetsma J.W. A prospective, community-based study on virologic assessment among elderly people with and without symptoms of acute respiratory infection. J. Clin. Epidemiol. 2003;56:1218–1223. doi: 10.1016/S0895-4356(03)00171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Mahajan S., Sidney J., Martini S., Scheuermann R.H., Peters B., Sette A. A survey of known immune epitopes in the enteroviruses strains associated with acute flaccid myelitis. Hum. Immunol. 2019;80:923–929. doi: 10.1016/j.humimm.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund E., Thomas A., Poore E.A., Amanna I.J., Rynko A.E., Mori M., Chen Z., Slifka M.K. Durability of vaccine-induced immunity Against tetanus and diphtheria toxins: A cross-sectional analysis. Clin. Infect. Dis. 2016;62:1111–1118. doi: 10.1093/cid/ciw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T., Rahman M., Ali F.A., Huang S.S.Y., Ata M., Zhang Q., Bastard P., Liu Z., Jouanguy E., Béziat V., et al. Distinct antibody repertoires against endemic human coronaviruses in children and adults. JCI Insight. 2021;6:e144499. doi: 10.1172/jci.insight.144499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killerby M.E., Biggs H.M., Haynes A., Dahl R.M., Mustaquim D., Gerber S.I., Watson J.T. Human coronavirus circulation in the United States 2014–2017. J. Clin. Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu R., Narean J.S., Wang L., Fenn J., Pillay T., Fernandez N.D., Conibear E., Koycheva A., Davies M., Tolosa-Wright M., et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat. Commun. 2022;13:80. doi: 10.1038/s41467-021-27674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang X., Nair H. Global seasonality of human seasonal coronaviruses: A clue for postpandemic circulating season of severe acute respiratory syndrome coronavirus. J Infect Dis. 2020;222:1090–1097. doi: 10.1093/infdis/jiaa436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.Y., Wolf J., Brice D.C., Sun Y., Locke M., Cherry S., Castellaw A.H., Wehenkel M., Crawford J.C., Zarnitsyna V.I., et al. Pre-existing humoral immunity to human common cold coronaviruses negatively impacts the protective SARS-CoV-2 antibody response. Cell Host Microbe. 2022;30:83–96.e4. doi: 10.1016/j.chom.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J.S., Vaqueirinho D., Mele F., Foglierini M., Jerak J., Perotti M., Jarrossay D., Jovic S., Perez L., Cacciatore R., et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science. 2021;372:1336–1341. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyal L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., et al. Cross-reactive CD4+ T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374:eabh1823. doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallajosyula V., Ganjavi C., Chakraborty S., McSween A.M., Pavlovitch-Bedzyk A.J., Wilhelmy J., Nau A., Manohar M., Nadeau K.C., Davis M.M. CD8+ T cells specific for conserved coronavirus epitopes correlate with milder disease in COVID-19 patients. Sci. Immunol. 2021;6:eabg5669. doi: 10.1126/sciimmunol.abg5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Dan J.M., Zhang Z., Rydyznski Moderbacher C., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickbakhsh S., Ho A., Marques D.F.P., McMenamin J., Gunson R.N., Murcia P.R. Epidemiology of seasonal coronaviruses: establishing the context for the emergence of coronavirus disease 2019. J. Infect. Dis. 2020;222:17–25. doi: 10.1093/infdis/jiaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega N., Ribes M., Vidal M., Rubio R., Aguilar R., Williams S., Barrios D., Alonso S., Hernández-Luis P., Mitchell R.A., et al. Seven-month kinetics of SARS-CoV-2 antibodies and role of pre-existing antibodies to human coronaviruses. Nat. Commun. 2021;12:4740. doi: 10.1038/s41467-021-24979-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee Y., Michelow I.C., Choe Y.J. Global seasonality of human coronaviruses: A systematic review. Open Forum Infect. Dis. 2020;7:ofaa443. doi: 10.1093/ofid/ofaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pene F., Merlat A., Vabret A., Rozenberg F., Buzyn A., Dreyfus F., Cariou A., Freymuth F., Lebon P. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin. Infect. Dis. 2003;37:929–932. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie J.G., Bazzi L.A., McDermott A.B., Follmann D., Esposito D., Hatcher C., Mateja A., Narpala S.R., O'Connell S.E., Martin E.T., Monto A.S. Coronavirus occurrence in the household influenza vaccine evaluation (HIVE) cohort of Michigan households: reinfection frequency and serologic responses to seasonal and severe acute respiratory syndrome coronaviruses. J. Infect. Dis. 2021;224:49–59. doi: 10.1093/infdis/jiab161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucinski S.L., Binnicker M.J., Thomas A.S., Patel R. Seasonality of coronavirus 229E, HKU1, NL63, and OC43 From 2014 to 2020. Mayo. Clin. Proc. 2020;95:1701–1703. doi: 10.1016/j.mayocp.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131:e143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletti G., Gerlach T., Jansen J.M., Molle A., Elbahesh H., Ludlow M., Li W., Bosch B.J., Osterhaus A.D.M.E., Rimmelzwaan G.F. Older adults lack SARS CoV-2 cross-reactive T lymphocytes directed to human coronaviruses OC43 and NL63. Sci. Rep. 2020;10:21447. doi: 10.1038/s41598-020-78506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva K.J., van de Sandt C.E., Lemke M.M., Lee C.Y., Shoffner S.K., Chua B.Y., Davis S.K., Nguyen T.H.O., Rowntree L.C., Hensen L., et al. Systems serology detects functionally distinct coronavirus antibody features in children and elderly. Nat. Commun. 2021;12:2037. doi: 10.1038/s41467-021-22236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggins M.K., Thwaites R.S., Openshaw P.J.M. Durability of immunity to SARS-CoV-2 and other respiratory viruses. Trends Microbiol. 2021;29:648–662. doi: 10.1016/j.tim.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Li W., Jiang S. Developing pan-beta-coronavirus vaccines against emerging SARS-CoV-2 variants of concern. Trends Immunol. 2022;43:170–172. doi: 10.1016/j.it.2022.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H.X., Lee W.S., Wragg K.M., Nelson C., Esterbauer R., Kelly H.G., Amarasena T., Jones R., Starkey G., Wang B.Z., et al. Adaptive immunity to human coronaviruses is widespread but low in magnitude. Clin. Transl. Immunology. 2021;10:e1264. doi: 10.1002/cti2.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell. Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend J.P., Hassler H.B., Wang Z., Miura S., Singh J., Kumar S., Ruddle N.H., Galvani A.P., Dornburg A. The durability of immunity against reinfection by SARS-CoV-2: a comparative evolutionary study. Lancet Microbe. 2021;2:e666–e675. doi: 10.1016/S2666-5247(21)00219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voic H., de Vries R.D., Sidney J., Rubiro P., Moore E., Phillips E., Mallal S., Schwan B., Weiskopf D., Sette A., Grifoni A. Identification and characterization of CD4+ T cell epitopes after Shingrix vaccination. J. Virol. 2020;94 doi: 10.1128/JVI.01641-20. e01641-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Shin J.H., Falsey A.R. Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. J. Infect. Dis. 2013;208:1634–1642. doi: 10.1093/infdis/jit393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Grunewald M., Perlman S. Coronaviruses: an updated overview of their replication and pathogenesis. Methods Mol. Biol. 2020;2203:1–29. doi: 10.1007/978-1-0716-0900-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterlow N.R., van Leeuwen E., Davies N.G., CMMID COVID-19 Working Group. Flasche S., Eggo R.M. How immunity from and interaction with seasonal coronaviruses can shape SARS-CoV-2 epidemiology. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2108395118. e2108395118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemeskel B.A., Dykema A.G., Garliss C.C., Cherfils S., Smith K.N., Blankson J.N. CD4+ T cells from COVID-19 mRNA vaccine recipients recognize a conserved epitope present in diverse coronaviruses. J. Clin. Invest. 2022;132:e156083. doi: 10.1172/JCI156083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wratil P.R., Schmacke N.A., Karakoc B., Dulovic A., Junker D., Becker M., Rothbauer U., Osterman A., Spaeth P.M., Ruhle A., et al. Evidence for increased SARS-CoV-2 susceptibility and COVID-19 severity related to pre-existing immunity to seasonal coronaviruses. Cell Rep. 2021;37:110169. doi: 10.1016/j.celrep.2021.110169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu E.D., Wang E., Garrigan E., Goodwin B., Sutherland A., Tarke A., Chang J., Gálvez R.I., Mateus J., Ramirez S.I., et al. Development of a T cell-based immunodiagnostic system to effectively distinguish SARS-CoV-2 infection and COVID-19 vaccination status. Cell Host Microbe. 2022;30:388–399.e3. doi: 10.1016/j.chom.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper uses publicly available datasets for epitope synthesis. Their references are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/k76f5sjjmh.1 Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request