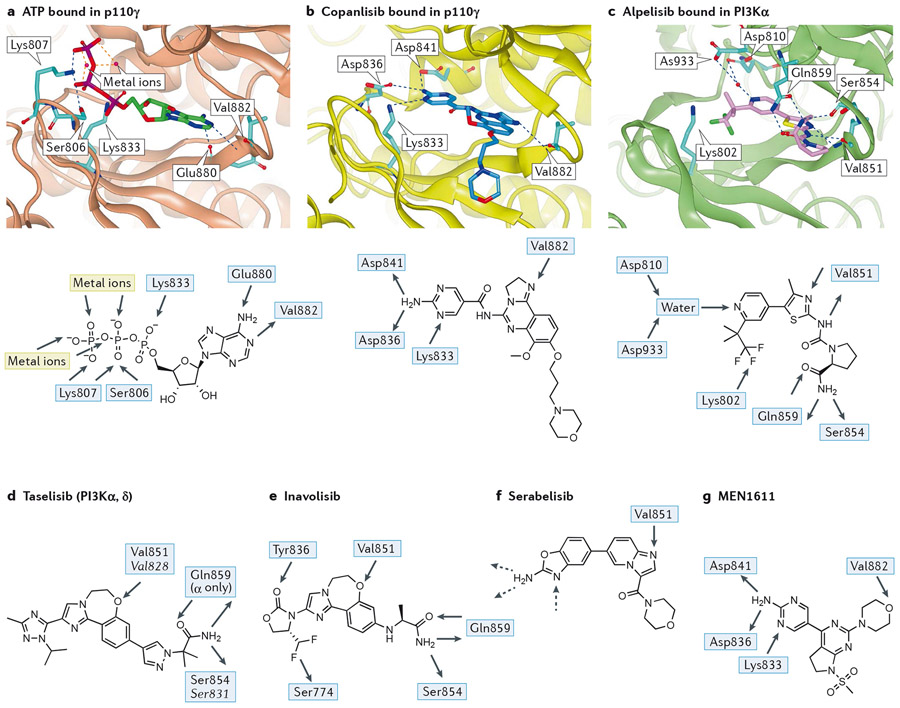
Figure 2 – Key features of the interaction between PI3Ks and pan- and PI3Kα-selective inhibitors.
The native shape of PI3K enzymes is taken to be that observed by crystallography for ATP-bound p110γ (2a; PDB:1E8X)18 or the very similar apo forms observed for p110γ (PDB:1E8Y)18, p110δ (PDB:2WXR)19 and PI3Kα (in complex with a partial p85α fragment, PBD:2RD0)20. Peptides are shown as ribbons with key residues shown in stick representation. Ligands are shown in stick representation. Colour coding of atoms in stick representations - carbon: cyan, oxygen: red; nitrogen: blue; fluorine: green; phosphorus: purple; colour coding of ligands - ATP: green, copanlisib: dark blue, alpelisib: pink, idelalisib: red; hydrogen bonds are shown in blue dashed lines, metal interactions in orange dashed lines.
a) (Top panel): ATP (carbon atoms bright green) bound in p110γ (1E8X)18; p110γ shown as brown ribbon with sidechains shown in cyan for residues mentioned in text. The adenine makes an acceptor-donor pair of hydrogen bonds with the NH of hinge Val882 and carbonyl of Glu880 whilst the triphosphate is bound by two metal ions, the terminal ammonium groups of Lys807 and Lys833 and a hydrogen bond from Ser806.(Lower panel): 2D representation of the interactions of ATP with the binding pocket of p110γ.
b) (Top panel) copanlisib (dark blue) bound in p110γ (yellow ribbon, 5G2N)23. The pendant aminopyrimidine group of copanlisib fits into the affinity pocket and forms H-bonds with Asp836 and Asp841 via the amino group and receives a H-bond from Lys833 to one of the ring nitrogen atoms. The morpholinopropyl moiety extends towards solvent and does not make any significant interactions; its role in the molecule is mainly as a solubilising group. (Lower panel): 2D representation of copanlisib indicating the H-bonds made with PI3Kγ.
c) (Top panel): alpelisib (pink) bound in PI3Kα (green ribbon, 4JPS)24. Note the multiple H-bonds: to hinge Val851; involving the primary carboxamide of alpelisib with Gln859 in p110α (Asp862, Lys890, Asn836 in p110β, γ and δ, respectively) and the backbone carbonyl of Ser854; the water-mediated H-bond to the pyridine N from Asp810 and Asp933. The charged terminal amine of Lys802 is close to the CF3 group. (Lower panel): 2D representation of the major interactions of alpelisib in PI3Kα.
d) 2D representation of the PI3Kα/δ inhibitor taselisib with H-bonding interactions observed in the crystal with PI3Kα and, in italic, with PI3Kδ. The ether oxygen of taselisib makes the key hinge interaction with both PI3Kα and PI3Kδ. Taselisib has a primary amide that can make the same interactions with p110α as alpelisib25, but in p110δ a rotation of the side chain places this amide differently, where it can still interact with the backbone carbonyl of Ser831 and places the terminal carbonyl of taselisib towards solvent (PDB:5T8F)360. In the affinity pocket, taselisib appears to be capapble of accepting H-bonds from Lys779 (PI3Kδ numbering) to N2 and from a putative water molecule located between Asp787 and Tyr813 (PI3Kδ numbering) to N4.
e) 2D representation of the PI3Kα-selective inhibitor inavolisib with H-bonding interactions observed in the crystal with PI3Kα. A carbonyl group in inavolisib accepts an H-bond from Tyr836 in p110α and a difluoromethyl group interacts with the hydroxyl of Ser774 in p110α. Although both of these residues are conserved in all class I PI3K isoforms, the combination of these structural features with a primary amide interacting with the non-conserved Gln859 of p110α results in very high PI3Kα isoform selectivity25.
f) Structure of the PI3Kα-selective inhibitor serabelisib. Although a crystal structure has not been disclosed for this molecule it is probable that the binding mode mimics that of copanlisib (Figure 2b) with the nitrogen of the imidazopyridine accepting a H-bond from the hinge Val851 and the aminobenzoxazole making interactions with the residues in the affinity pocket (hashed arrows).
g) Structure of PI3Kα inhibitor MEN1611 showing the observed hydrogen bonds in PI3Kγ.