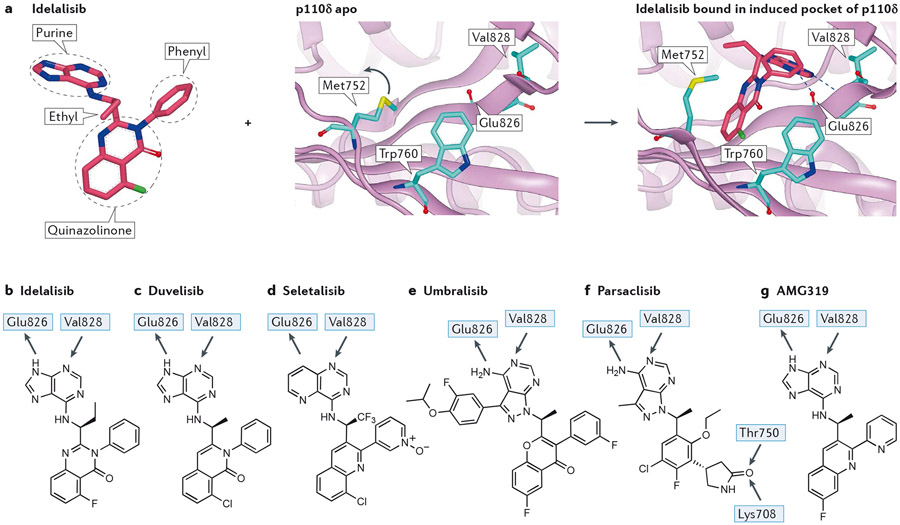
Figure 4 – Interactions of selected propeller-shaped PI3Kδ-selective inhibitors with PI3Kδ.
a) Inhibitor-induced specificity pocket in PI3K, illustrated by idelalisib binding to p110δ. Left panel: structure of idelalisib from 4XEO drawn to emphasise the propeller shape, thus the three ring systems of the hinge-binding purine, the quinazolinone amd the phenyl are approximately mutually orthogonal in an orientation organised by a combination of the chiral ethyl group and the phenyl ring. Middle panel: apo structure of p110δ (2WXR) with Met752 packing against Trp760. The blue arrow indicates the relative motion of Met752 in the flexing of the enzyme in solution that can open up the selectivity pocket. Right panel: crystal structure of idelalisib bound in p110δ (4XEO) with the purine making the hinge interaction with the NH of Val828 and the carbonyl of Glu826. The electron deficient quinazolinone ring system fits into the induced selectivity pocket between Met752 and Trp760 and makes a face to edge interaction with the electron rich indole of Trp760.
b) 2D representation of idelalisib showing the major interactions with p110δ.
c) 2D representation of the PI3Kγ/δ inhibitor duvelisib, with H-bonding interactions observed in the crystal with p110δ. Note the similarity to idelalisib.
d) 2D representation of the PI3Kδ-selective inhibitor seletalisib. This is another propeller-shaped PI3Kδ inhibitor, in this case it is probable that the 1 N atom accepts an H-bond from the hinge Val828, with a non-classical H-bond being formed from the CH of the adjacent pyridine ring.
e) Structure of PI3Kδ/CK 1ε inhibitor umbralisib. A crystal structure of this has not been published; however, based on the similarity with other propeller inhibitors the structural features can be identified with confidence. The 3-fluoro-4-isopropoxyphenyl ring is similar to substituents in SW13 and SW14 for which crystal structures are known19; this occupies the affinity pocket and may be responsible for the high isoform selectivity observed.
f) Structure of the PI3Kδ-selective inhibitor parsaclisib with proposed H-bonding interactions based on molecular docking. Note the additional interactions made by the pendant lactam that accepts two H-bonds from both the hydroxyl of Thr750 (p110δ, Arg770, Lys777, Lys802 in p110α, β and γ, respectively) and the terminal ammonium of Lys708 (p110δ, Gln728, Arg735, Ser760 in p110α, β and γ), respectively; other propeller inhibitors do not have an equivalent group. Despite the multiple structural differences with other PI3Kδ inhibitors, parsaclisib still forms a propeller shape.
g) Structure of PI3Kδ-selective inhibitor AMG319 showing the hinge interactions with PI3Kδ based on a crystal structure in PI3Kγ.