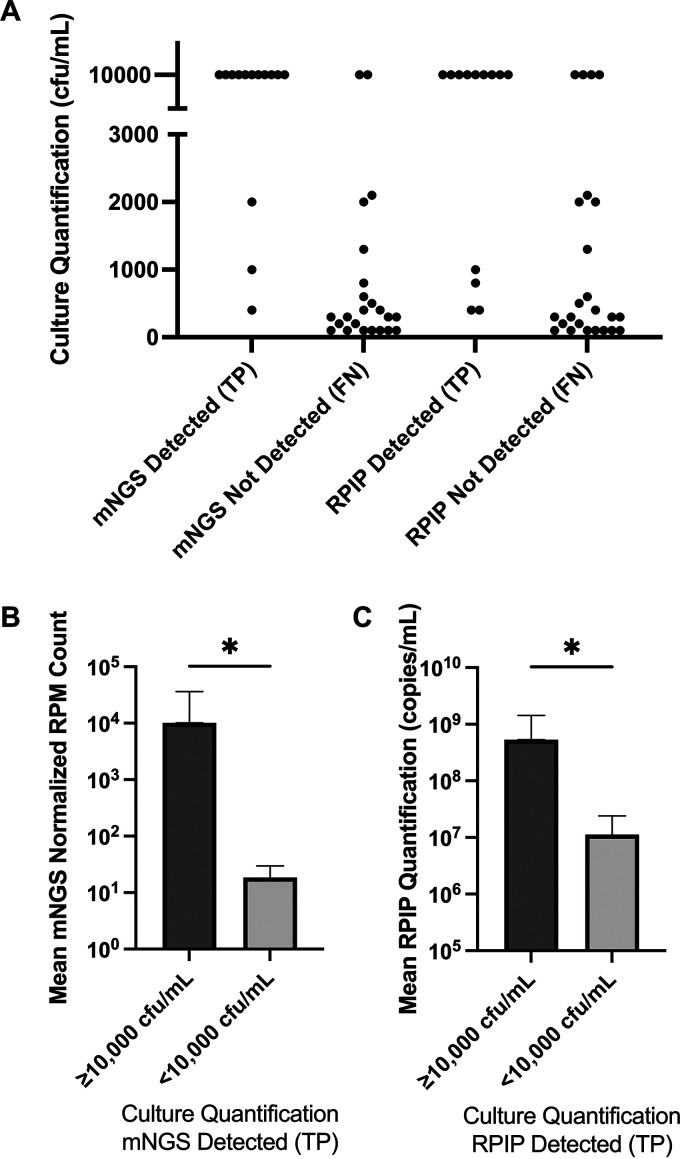
FIG 3.
Relationship of bacteria quantified by standard methods and those detected by NGS workflows. (A) True-positive (TP) and false-negative (FN) results per workflow. Each data point represents bacteria isolated and quantified from standard aerobic cultures (n = 37). Isolates reported as ≥10,000 CFU/mL were plotted at 10,000 CFU/mL. Bacteria detected by standard culture with semiquantification or without quantification were not included. The metagenomic NGS (mNGS) workflow detected 11 of 13 isolates (84.6%) quantified at >10,000 CFU/mL but did not detect 21 of 24 isolates (87.5%) quantified at <10,000 CFU/mL. Similarly, the RPIP targeted workflow detected 9 of 13 isolates (69.2%) quantified at >10,000 CFU/mL but did not detect 20 of 24 isolates (83.3%) quantified at <10,000 CFU/mL. (B and C) Relationship of NGS quantification methods to relative culture abundance for true-positive samples. Statistical comparisons were made using Mann-Whitney testing (P = 0.02 for mNGS, and P = 0.03 for RPIP targeted NGS). Error bars represent standard deviations. Note the difference in the y axes.