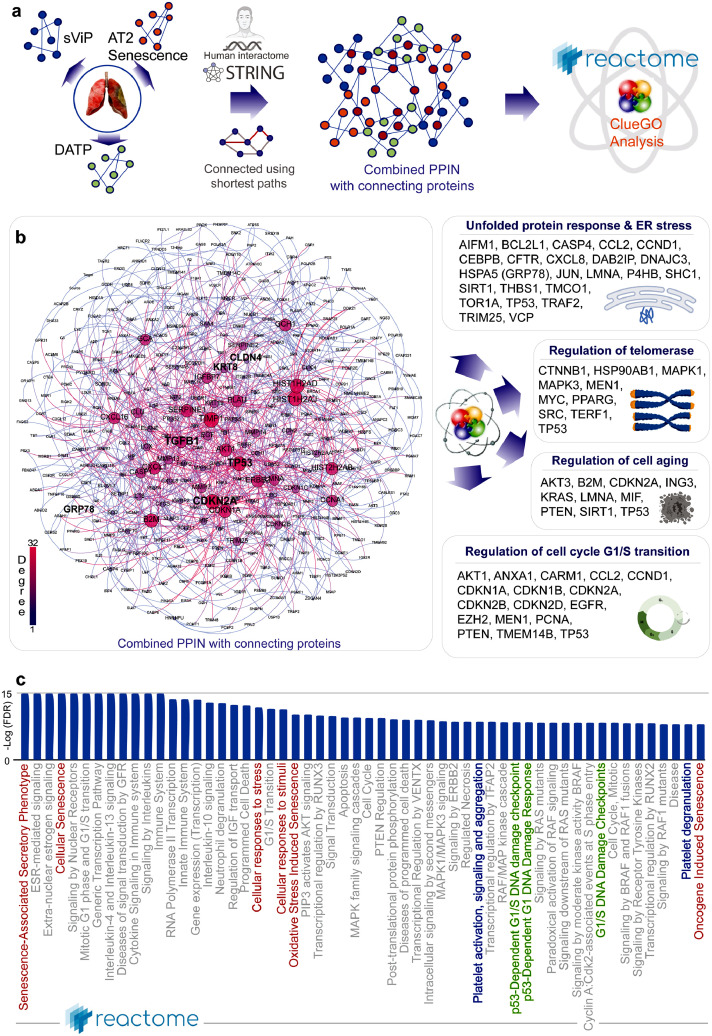
Figure 7.
Protein-protein interaction network (PPIN) analysis predicts ER stress in AT2 cells as a driver pathophysiology in COVID-19. a. Schematic summarizes the key steps in building a PPI network using as ‘seeds’ the genes in the sVIP, AT2 senescence and transition state arrest (DATP) signatures and connecting them using shortest paths prior to carrying out network analyses. b. The combined PPI network is shown on the left and the degree of connectivity is indicated. See also Supplemental Information 5 for the complete list of nodes sorted by their degrees of connectivity. Schematics on the right summarize the results of CluGo analysis, which identifies cellular processes and key genes that may perpetuate lung fibrosis. c. Reactome pathway analysis of the PPI network lists the most significant pathways that are associated with the node list in the network in B. Red = senescence related processes; Green = DNA damage response pathways; Blue = Platelet activation and degranulation.