FIGURE 1.
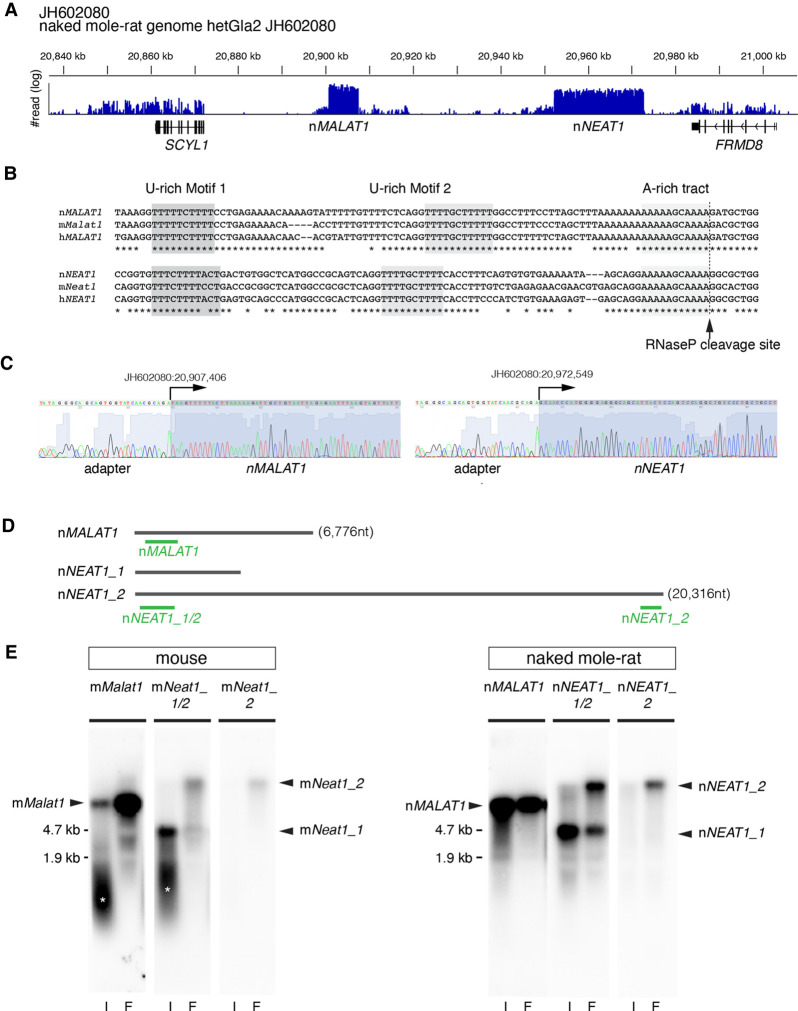
Identification of MALAT1 and NEAT1 in naked mole-rats. (A) Distribution of RNA-seq reads mapped to the region of the naked mole-rat genome syntenic to mouse chromosome 19 containing mMalat1 (mouse Malat1) and mNeat1 (mouse Neat1), which are flanked by SCYL1 and FRMD8. Note that two regions were abundantly transcribed into RNA, which corresponded to nMALAT1 (naked mole-rat MALAT1) and nNEAT1 (naked mole-rat NEAT1), respectively. (B) Sequence alignment of the 3′ regions of NEAT1 and MALAT1 in naked mole-rats (n), mouse (m), and human (h). The tandem U-rich motifs followed by A-rich tract, which form triple helix structures, are highlighted in shadow boxes and are highly conserved between the three species. The arrow indicates the putative RNaseP cleavage site that generates the 3′ end of these transcripts. (C) Sequence analyses of PCR fragments obtained by the 5′ RACE analyses. Arrows indicate the positions of the transcription start sites of nMALAT1 and nNEAT1. (D) Schematic drawing indicating the positions of probes used for northern blot and in situ hybridization studies. (E) Northern blot analyses of nMALAT1 and nNEAT1 expression in embryonic fibroblast cells and intestine. (I) intestine, (F) fibroblasts. Asterisks show the signals derived from degraded RNAs.