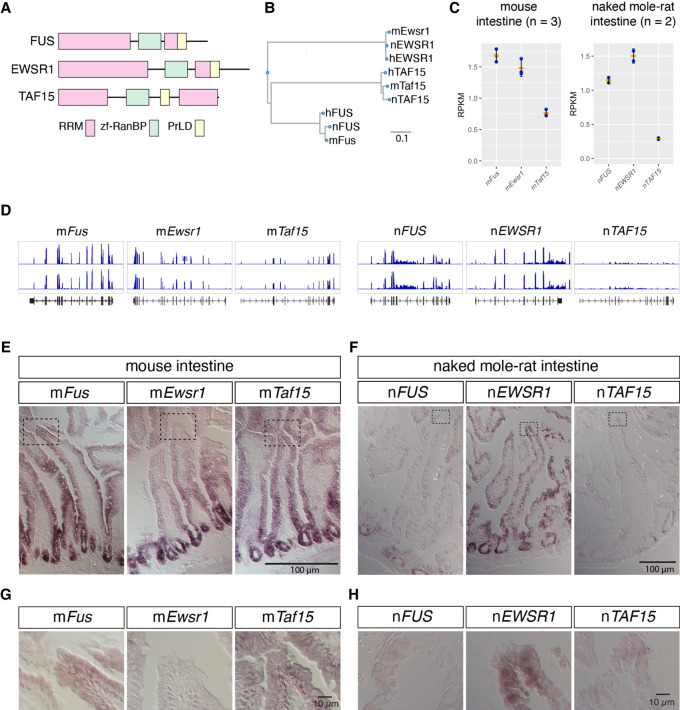
FIGURE 7.
mRNAs of FET family proteins are redundantly expressed in intestine of naked mole-rats. (A) Domain structures of FET family proteins. PrLD: prion like domain, RRM: RNA recognition motif, zf-RanBP: zinc finger motif in Ran binding protein. The PrLD were predicted using PLAAC (Lancaster et al. 2014). (B) Phylogenetic tree of FET family proteins of human (h), mouse (m), and naked mole-rat (n). Scale bar indicates the amino acid substitution ratio per residue. (C) Quantification of gene expression of FET family proteins. The expression was estimated by RNA-seq analyses for mouse intestine and NS-fibroblasts. RPKM: reads per kilobase of transcript per million mapped reads. Note the higher expression of nEWSR1 compared to nFUS in naked mole-rats. Orange bars represent the mean value, blue dots represent the value of each sample, and black bars represent standard deviations. (D) Distribution of RNA-seq reads (blue) mapped to FET family genes in the genome of mice (mm10) and naked mole-rats (hetGla2). Gene structures are shown in black color in the bottom track. (E–H) In situ hybridization analyses of the expression of FET family proteins in the intestine of mice (E,G) and naked mole-rats (F,H). Dashed boxes in E and F indicate the regions shown at a higher magnification in G and H. Note that nEWSR1 was expressed in the distal tip cells of the intestinal epithelium of naked mole-rats. Scale bars, 100 µm in E and F, and 10 µm in G and H.