Fig. 6.
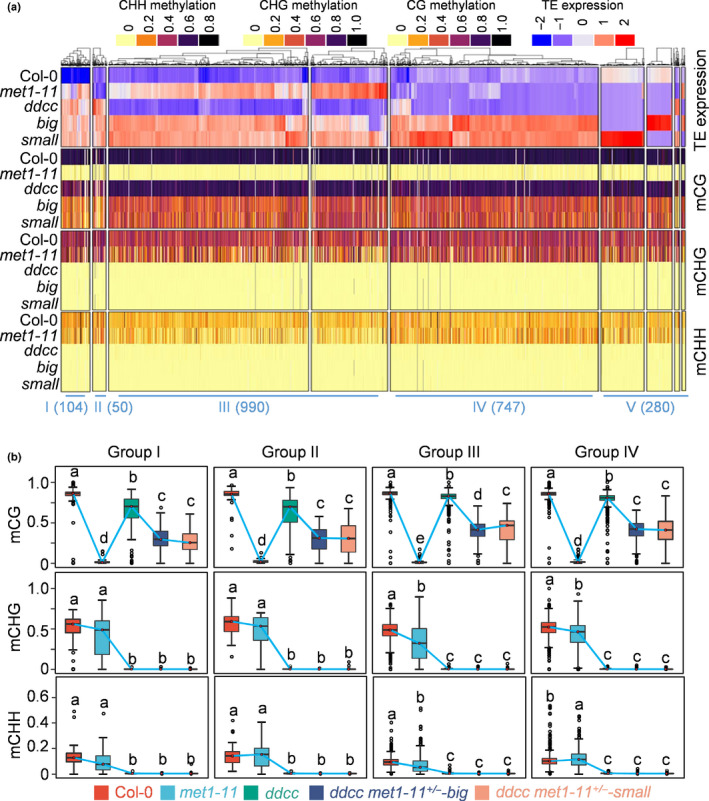
CG and non‐CG DNA methylation synergistically and redundantly control transposable element (TE) silencing in Arabidopsis. (a) Heat map of the activated TEs in the ddcc met1‐11+/− mutant plants. The DNA methylation levels of each TE are shown. These activated TEs are divided into five groups. Group I, TEs that are activated in either met1 or ddcc. Group II, TEs that are activated in ddcc but not in met1. Group III, TEs that are activated in met1 but not in ddcc. Group IV, TEs that remain silenced in either met1 or ddcc, but are activated in ddcc met1‐11+/− . Group V, others TEs. The number of TEs in each group is shown. big, ddcc met1‐11+/−‐big; small, ddcc met1‐11+/−‐small. (b) Box plots show the DNA methylation levels of the four groups of TEs in the indicated mutants. Dark horizontal line, median; edges of boxes, 25th (bottom) and 75th (top) percentiles; whiskers, minimum and maximum percentage of DNA methylation. Significant differences between two groups are marked with different letters (P < 10−15, Mann–Whitney U‐test).
