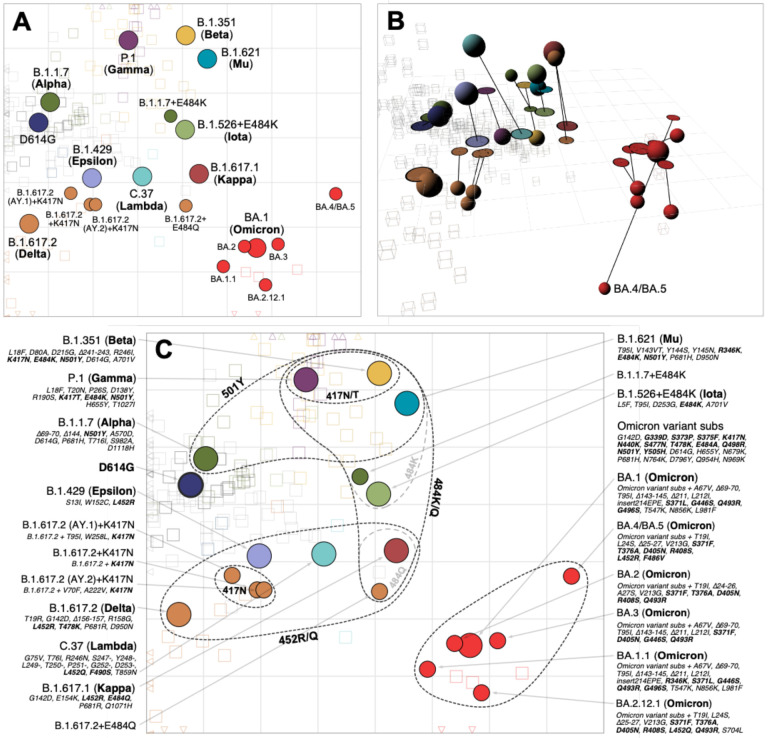
Fig. 3. Antigenic map of SARS-CoV-2 variants and selected substitutions.
A) Antigenic map with variant names. B) Antigenic map with variant positions in 3D and lines connecting to their respective positions in the 2D map. C) Antigenic map with variant names and substitutions annotated and grouped by amino acid present at spike positions 417, 452, 484 and 501, with an additional grouping for the 6 Omicron variants. Variants are shown as circles, sera as squares/cubes. Variants with additional substitutions from a root variant are denoted by smaller circles, in the color of their root variant. The x and y-axes both represent antigenic distance, with one grid square corresponding to a two-fold serum dilution in the neutralization assay. Therefore, two grid squares correspond to a four-fold dilution, three to an eight-fold dilution and so on. The x-y orientation of the map is free, as only the relative distances between variants and sera are relevant. Triangular arrowheads at the edge of the bounding box point in the direction of the sera that would be shown outside of the plot limits. A non-zoomed version of this map is shown in fig. S22. Interactive versions of the maps shown in panels A & B are available online at https://acorg.github.io/mapping_SARS-CoV-2_antigenic_relationships_and_serological_responses.