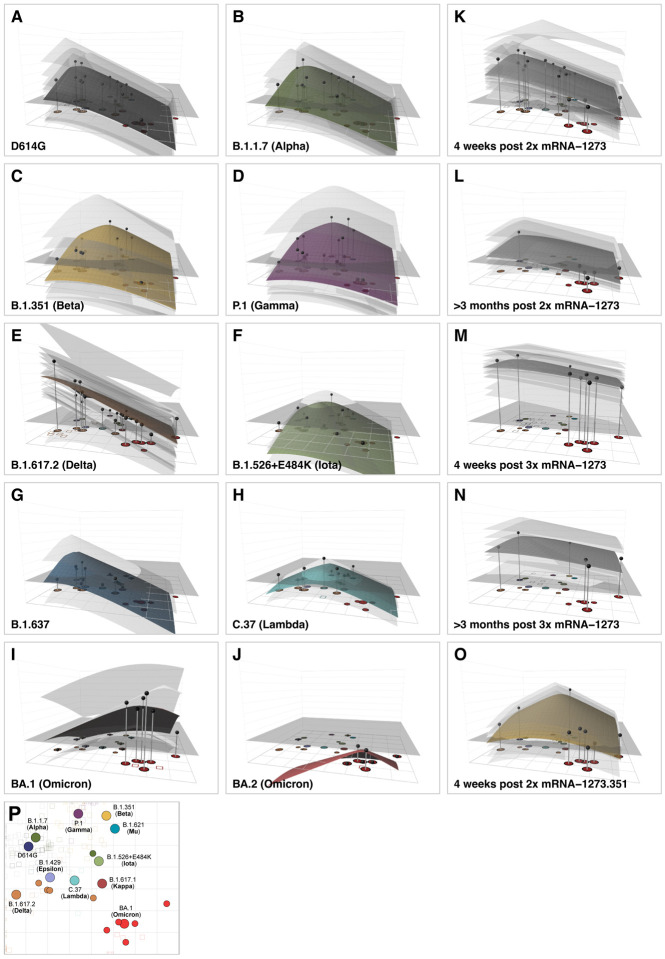
Fig. 4. Antibody landscapes for each serum group.
Colored surfaces show the GMT antibody landscapes for the different serum groups, light gray surfaces show the landscapes for individual sera. Gray impulses show the height of the GMT for a specific variant, after accounting for individual effects as described in Materials and Methods (which would otherwise bias the GMT for variants not titrated against all sera). The base x-y plane corresponds to the antigenic map shown in Fig. 3 and reproduced in panel P. The vertical z-axis in each plot corresponds to the titer on the log2 scale, each two-fold increment is marked, starting from a titer of 20, one unit above the map surface. The gray horizontal plane indicates the height of a titer of 50, as a reference for judging the landscapes against various estimates of neutralizing antibody correlates of protection. Additional visualizations of predicted versus fitted titers are shown in fig. S24. The number of sera included for the calculation of the landscapes are A) D614G sera (n=13), B) B.1.1.7 sera (n=13), C) B.1.351 sera (n=15), D) P.1 sera (n=13), E) B.1.617.2 sera (n=21), F) B.1.526+E484K sera (n=4), G) B.1.637 sera (n=2), H) C.37 sera (n=2), I) BA.1 sera (n=4), J) BA.2 sera (n=1), K) 4 weeks post 2x mRNA-1273 sera (n=30), L) >3 months post 2x mRNA-1273 sera (n=13), M) 4 weeks post 3x mRNA-1273 sera (n=26), N) >3 months post 3x mRNA-1273 sera (n=8), O) 4 weeks post 2x mRNA-1273.351 sera (n=9). Interactive versions of the landscapes in each of the panels are available online at https://acorg.github.io/mapping_SARS-CoV-2_antigenic_relationships_and_serological_responses.