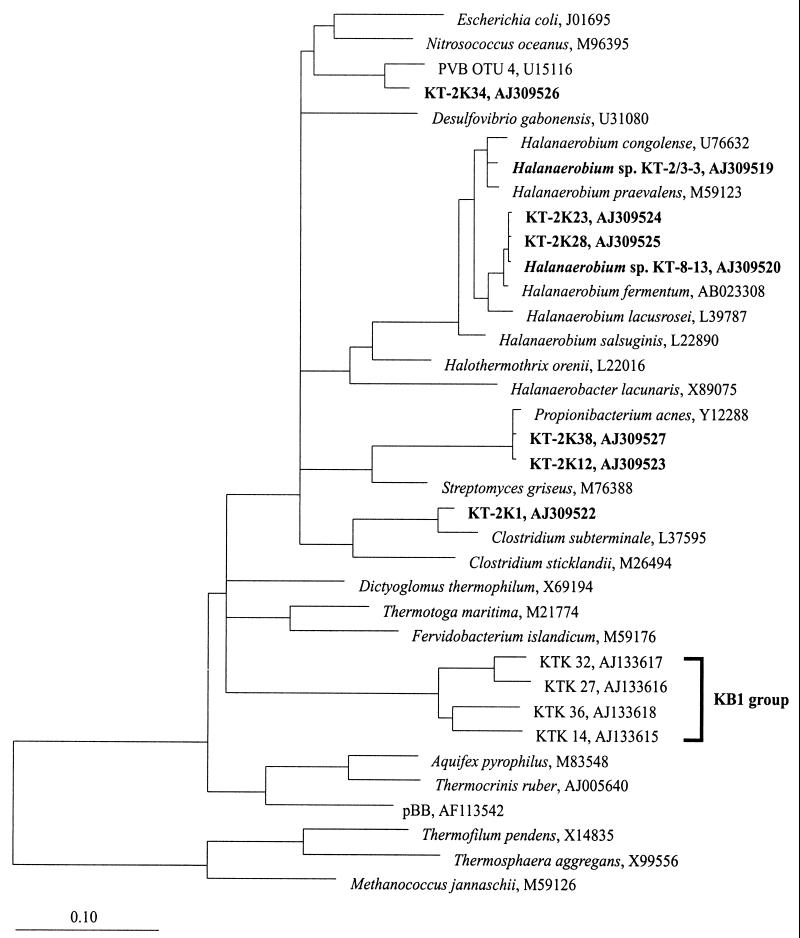
FIG. 5.
16S rRNA gene-based phylogenetic tree of the bacterial domain, including the 16S rDNA sequences from brine-seawater interface sample KT-2 and the 16S rRNA gene sequences of the new halophilic isolates from Kebrit Deep, Red Sea. The KB1 group marks a cluster of closely related environmental sequences, which were obtained from a sediment sample (depth, 1,515 m) of the Kebrit Deep (21). The topology of the tree is based on results of a maximum-parsimony analysis. Reference sequences were chosen to represent the broadest diversity of Bacteria. Only sequence positions that share 50% or more residues identical to the 16S rRNA sequences of a corresponding group were included for tree reconstruction. Accession numbers for the sequences are indicated. The scale bar represents 0.10 fixed mutation per nucleotide position.