Figure 6.
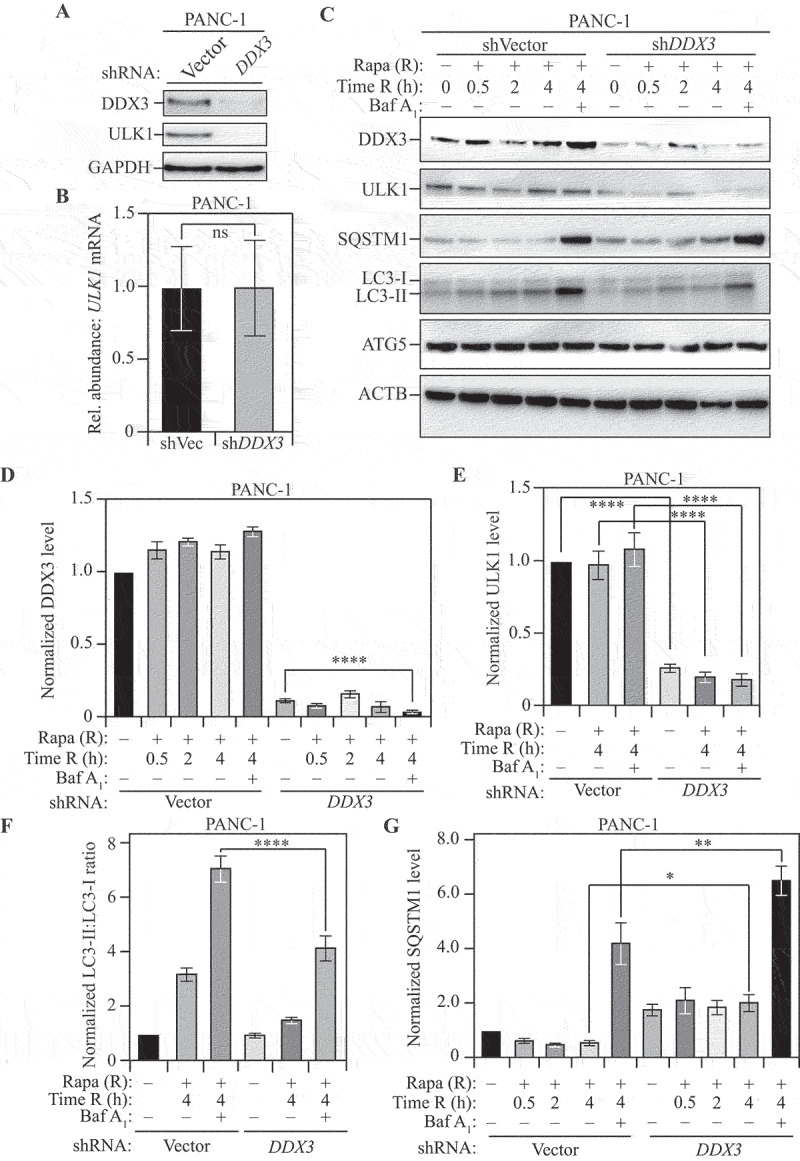
DDX3 regulates autophagy in mammalian cells. (A) Stable shRNA-mediated knockdown of DDX3 in PANC-1 cells: Western blotting to probe for DDX3 and ULK1 levels in cells transfected with shVector (control) or shDDX3. GAPDH was used as a loading control. (B) Relative abundance of ULK1 mRNA in shDDX3 cells compared to control cells. GAPDH was used as a reference gene. Data represent three independent biological replicates. (C) The loss of DDX3 leads to a reduction in ULK1 levels and autophagy in mammalian cells: PANC-1 cells, stably transfected with either vector shRNA (control) or shRNA targeting DDX3, were treated with rapamycin (Rapa; R) for the indicated times with or without co-treatment with bafilomycin A1 (Baf A1). A representative blot shows the levels of DDX3, ULK1, SQSTM1, LC3-I, LC3-II, and ATG5 with ACTB as a loading control, upon harvesting cells at the indicated time points after rapamycin treatment. (D and E) Normalized DDX3 (D) and ULK1 (E) levels at the indicated time points with the indicated treatments. Data represent three independent biological replicates. (F) Normalized LC3-II:LC3-I ratio at the indicated time points with the indicated treatments. Decreased LC3-II:LC3-I ratio in the presence of bafilomycin A1 indicates reduced autophagy flux. Data represent three independent biological replicates. (G) Normalized SQSTM1 level at the indicated time points with the indicated treatments. Increased SQSTM1 accumulation indicates reduced autophagy flux. Data represent three independent biological replicates. Data in (B) and (D-G) represent mean ± SEM from indicated number of replicates. (B) was analyzed using unpaired Student’s t-test whereas the statistical analysis for (D-G) was carried out using one-way analysis of variance (ANOVA). Multiple comparisons were carried out using Tukey’s multiple comparisons test. *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001 ns: not significant.
