Abstract
Objective
To determine the frequency of germline and somatic pathogenic BRCA1 and BRCA2 variants in patients with high‐grade serous ovarian cancer tested by next‐generation sequencing (NGS), with the aim of defining the best strategy to be implemented in future routine testing.
Design
National retrospective audit.
Setting
The All Wales Medical Genomics Service (AWMGS).
Population
Patients with high‐grade serous ovarian/fallopian tube/peritoneal cancer referred by oncologists to the AWMGS between February 2015 and February 2021 for germline and/or tumour testing of the BRCA1 and BRCA2 genes by NGS.
Methods
Analysis of NGS data from germline and/or tumour testing.
Main outcome measures
Frequency of BRCA1 and BRCA2 pathogenic variants.
Results
The overall observed germline/somatic pathogenic variant detection rate was 11.6% in the 844 patients included in this study, with a 9.2% (73/791) germline pathogenic variant detection rate. Parallel tumour and germline testing was carried out for 169 patients and the overall pathogenic variant detection rate for this cohort was 14.8%, with 6.5% (11/169) shown to have a somatic pathogenic variant. Two BRCA1 dosage variants were found during germline screens, representing 2.0% (2/98) of patients with a pathogenic variant that would have been missed through tumour testing alone.
Conclusions
Parallel germline and tumour BRCA1 and BRCA2 testing maximises the detection of pathogenic variants in patients with high‐grade serous ovarian cancer.
Tweetable abstract
Parallel germline and tumour testing maximises BRCA pathogenic variant detection in ovarian cancer.
Keywords: BRCA, germline, mainstreamed, oncology‐led, ovarian cancer, pathogenic variant, somatic
Tweetable abstract
Parallel germline and tumour testing maximises BRCA pathogenic variant detection in ovarian cancer.
Linked article This article is commented on by C Gourley, p. 443 in this issue. To view this mini commentary visit https://doi.org/10.1111/1471-0528.16978.
Introduction
The identification of a pathogenic variant in either the BRCA1 or BRCA2 gene (hereafter collectively referred to as BRCA) can have prognostic and predictive implications for patients with ovarian cancer and their families. Poly (ADP‐ribose) polymerase inhibitor (PARPi) treatment significantly improves progression‐free survival in patients with advanced ovarian cancer, particularly in those with germline and somatic BRCA pathogenic variants. 1 , 2 , 3 , 4 , 5 , 6 Additionally, family members that are subsequently found to have a germline pathogenic BRCA variant through pre‐symptomatic cascade testing are at an increased risk of developing breast and/or ovarian cancer. 7 The identification of individuals with pathogenic BRCA variants is therefore crucial to be able to guide treatment and inform at‐risk individuals about appropriate clinical management strategies, which may involve increased surveillance and/or risk‐reducing procedures. 8
Recent UK studies estimate the prevalence of germline pathogenic BRCA variants at 7.8–18.5% in patients with various types of epithelial ovarian cancer (Table 1). 9 , 10 , 11 , 12 , 13 , 14 Studies in patients with serous ovarian cancer have shown that a further 4.1–7.1% of patients’ tumours harbour an acquired BRCA pathogenic variant (Table 2). 15 , 16 , 17 This highlights a significant proportion of this patient population that may be more responsive to PARPi maintenance therapy and demonstrates the clinical need to have effective genetic screening strategies in National Health Service (NHS) laboratories to identify these patients in a timely manner.
Table 1.
Comparison of germline testing results from this study with similar recent UK‐based studies
| This study | Rust, 2018 9 | Rahman, 2019 10 | Rumford, 2020 11 | George, 2016 12 | Plaskocinska, 2016 13 | Flaum, 2020 14 | |
|---|---|---|---|---|---|---|---|
| Number of patients | 791 | 236 | 122 | 255 | 207 | 232 | 480 |
| Data collection dates | 02/2015–02/2021 | 2013–NR | 02/2015–04/2016 | 04/2016–04/2018 | 07/2013–11/2014 | 07/2013–06/2015 | 11/2017–11/2019 |
| Setting | AWMGS, Wales | Four centres across Scotland | UCLH, England | Imperial College Hospital, England | The Royal Marsden Hospital, England | East Anglian Genetics Services, England | North West of England |
| Testing criteria | High‐grade serous ovarian/fallopian tube/peritoneal cancer | Non‐mucinous ovarian cancer | High‐grade non‐mucinous ovarian cancer | Epithelial ovarian cancer | Non‐mucinous ovarian cancer | Newly diagnosed epithelial ovarian cancer | Non‐mucinous epithelial cancer of ovary/fallopian tube/peritoneum |
| Age (years) at diagnosis (or referral where appropriate) of patients with versus without pathogenic variant | 58.1 vs 67.7 (mean) | NR | 58 vs 62 (median) | 57.8 vs 62.9 (mean) | 53.9 vs 57.8 (mean) | 49.5 vs 66.1 (mean) | 53.1/60.5 (BRCA1/BRCA2) vs 60.5 (mean) |
| Germline pathogenic BRCA variant detection rate | 73 (9.2%) | 31 (13.1%) | 18 (14.8%) | 34 (13.3%) | 33 (15.9%) | 18 (7.8%) | 89 (18.5%) |
| Dosage variant rate | 2 (2.7%) | NR | NR | 2 (5.9%) | 4 (12.1%) | NR | NR |
| Founder variant rate | 2 (2.7%) | NR | 2 (11%) | 6 (17.6%) | NR | 0 (0%) | AKJ population excluded |
AKJ, Ashkenazi Jewish; NR, not reported.
Categorical data presented as number of patients (%).
Table 2.
Comparison of parallel tumour and germline testing results from this study with similar recent studies
| This study | Koczkowska, 2016 15 | Eoh, 2020 16 | Peixoto, 2020 17 | |
|---|---|---|---|---|
| Number of patients | 169 | 97 | 98 | 95 |
| Setting | AWMGS, Wales | Medical University of Gdansk, Poland | Severance Hospital, South Korea | Portuguese Oncology Institute of Porto, Portugal |
| Testing criteria | High‐grade serous ovarian/fallopian tube/peritoneal cancer | Serous ovarian carcinoma | High‐grade serous ovarian cancer | High‐grade serous ovarian cancer |
| Overall (somatic and/or germline) pathogenic variant detection rate | 25 (14.8%) | 27 (27.8%) | 24 (24.5%) | 22 (23.2%) |
| Acquired pathogenic variant rate | 11 (6.5%) | 4 (4.1%) | 7 (7.1%) | 6 (6.3%) |
Data presented as number of patients (%).
BRCA genetic testing in NHS laboratories is currently performed through germline and/or tumour next‐generation sequencing (NGS). Acquired somatic variants can only be detected through tumour testing whereas germline variants can be detected by both testing methods. However, most NGS techniques currently employed by NHS laboratories to test tumour tissue are unable to detect large rearrangements and dosage variants. Since 2015, BRCA testing through the All Wales Medical Genomics Service (AWMGS) has been offered to all patients diagnosed with high‐grade serous ovarian/fallopian tube/peritoneal cancer in Wales through an oncologist‐led, mainstreamed referral pathway. 18 A concurrent dual‐testing strategy, where germline and tumour testing is requested in parallel, has been employed in Wales since 2019, in‐line with current British Gynaecological Cancer Society guidelines. 19 The purpose of this study is to audit the outcomes of these testing strategies to compare the detection of BRCA pathogenic variants. This is with the aim of determining the most effective strategy for the timely identification of women who may benefit most from PARPi maintenance therapy to adopt in future routine testing.
Methods
Patient population
This was a retrospective audit of the records of patients with high‐grade serous ovarian/fallopian tube/peritoneal cancer who were referred by oncologists across Wales to the AWMGS (Cardiff, Wales) between February 2015 and February 2021 for germline and/or tumour testing of the BRCA1 and BRCA2 genes by NGS. There was no patient/public involvement in the design of this audit. Consent for testing was obtained by the referring oncologist. Counselling and results were also delivered by the referring oncologist.
Evolution of the testing strategy
Germline BRCA NGS analysis for patients diagnosed with high‐grade serous ovarian/peritoneal/fallopian tube cancer through the mainstreamed, oncology‐led diagnostic referral pathway began in early 2015. The BRCA tumour testing pathway for the same patient population was launched in 2017. Initially tumour samples were only tested in relapsed patients with a normal germline result in a two‐step testing strategy. Since the start of 2019, it has been possible to request parallel tumour and germline tests concurrently for newly diagnosed patients. Figure 1 displays the key changes to the testing strategy in relation to the number of germline and tumour tests that were requested since the introduction of this novel, mainstreamed pathway in 2015.
Figure 1.
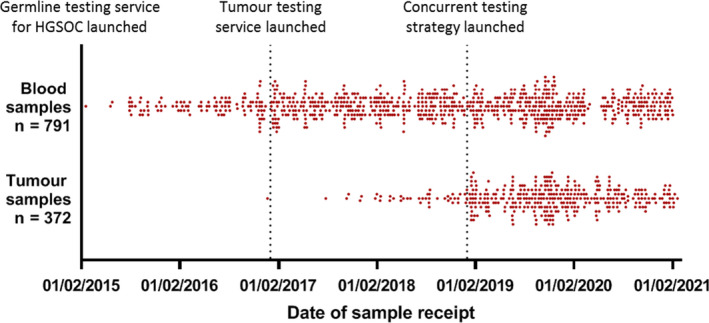
Distribution of dates of arrival of samples in AWMGS of blood and tumour samples referred for germline and tumour BRCA1/2 tests, respectively. HGSOC, high‐grade serous ovarian cancer.
Germline testing
DNA was extracted from blood samples. Analysis of the BRCA1 and BRCA2 genes was carried out using the Illumina TruSight® Cancer Sequencing Panel which uses the Illumina TruSight® Rapid Capture protocol for library preparation and target enrichment. Targeted gene NGS was performed using an Illumina® NextSeq 550 sequencing system using the Illumina® TG NextSeq 500/550 Mid Output Kit v2.5 (300 Cycles). The minimum read depth was 20× over the region of interest, which was coding exons ±5 flanking intronic bases (although some variants outside this region were also detectable if the minimum quality thresholds were met). A custom bioinformatics pipeline was used to call and annotate single nucleotide polymorphisms, insertions/deletions and dosage (whole exon/gene deletions/duplications) variants. Sanger sequencing or multiplex ligation‐dependent probe amplification where appropriate were used to confirm variants. Referral to the AWMGS clinical genetics service was recommended in cases where a germline pathogenic variant was identified.
Tumour testing
DNA was extracted from formalin‐fixed, paraffin‐embedded tumour samples with a minimum tumour nuclei content of 10%. Multiplex PCR‐based target enrichment of BRCA1 and BRCA2 was performed using the Qiagen GeneRead™ DNAseq Targeted Panels V2 kit and the NGHS‐102X primer panel (Qiagen, Hilden, Germany). NGS was carried out using an Illumina® MiSeq® sequencer (Illumina, San Diego, CA, USA). Minimum read depth over the region of interest (coding exons ±5 flanking intronic bases) was 500× unless the coverage at this depth was less than 80% for either BRCA1 or BRCA2, in which case the minimum read depth was 100× (as long as the tumour content of the sample was more than 50%). A custom bioinformatics pipeline was used to call and annotate single nucleotide polymorphisms and small insertions/deletions. This methodology does not allow the detection of dosage/structural variants and the sensitivity for deletions/insertions larger than a few bases is not well characterised. Tumour testing allows the detection of both germline and acquired somatic variants but it is not possible to distinguish between these without carrying out a paired germline analysis. Where a pathogenic variant was identified in the tumour, germline testing was recommended to investigate the possibility of germline origin.
Variant analysis
Variants were analysed using the current American College of Medical Genetics and Genomics (ACMG) and Association for Clinical Genomic Science (ACGS) variant classification guidelines. 20 , 21 The Cancer Variant Interpretation Group UK (CanVIG‐UK) specifications for the interpretation of germline variants in cancer susceptibility genes were also applied. 22 Reference sequences: NM_007294.3 (BRCA1) and NM_000059.3 (BRCA2). Pathogenic variants in this study refer to those classified as ‘likely pathogenic’ (class 4) or ‘pathogenic’ (class 5) according to ACMG guidelines. Variants classified as a variant of uncertain significance are not clinically actionable in terms of guiding treatment pathways for this patient population and have not been reported in this audit.
Statistical analysis
Categorical data are described using counts and percentages. For the percentages of patients with a pathogenic variant, 95% confidence intervals were calculated. Simple descriptive statistics (mean, standard deviation [SD]) were computed for the age at referral of patients and the range and the time between referrals. GraphPad Prism 8 was used to produce graphs (GraphPad, San Diego, CA, USA).
Results
Testing pathways
Data were available for a total of 844 women (mean age at referral 67.0 years, SD 10.2) who underwent a germline and/or tumour BRCA test. A total of 472 patients only received germline testing and 53 patients only received tumour testing. The remaining 319 patients received dual testing (both a germline and a tumour test) with a subset of these (n = 169) receiving dual testing in parallel, where testing of the tumour sample and the germline sample for that individual overlapped and the referral into each testing pathway was therefore independent from the outcome of the other test. Figure 2 and Table 3 summarise the number of patients according to the testing strategy they received.
Figure 2.
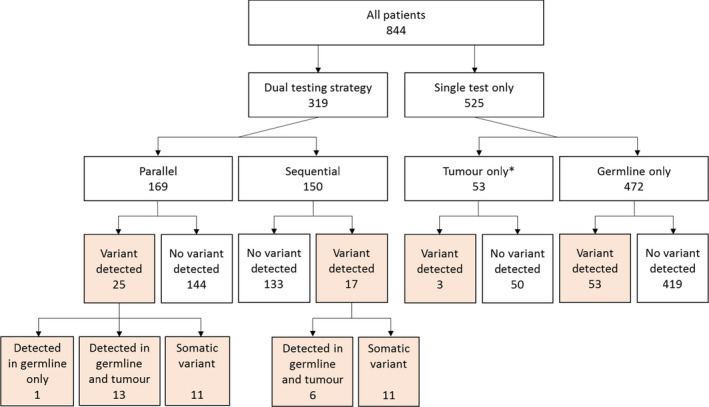
Flow diagram detailing the number of patients according to testing received and outcome (pathogenic variant detection). *Please note: testing of a tumour sample only is not a standalone testing strategy. It is anticipated that further germline testing will be carried out for the majority of these patients.
Table 3.
Prevalence of pathogenic BRCA1/2 variants according to testing strategy
| Germline testing* | Tumour testing* | Dual testing | All patients included in study, regardless of testing strategy | |||
|---|---|---|---|---|---|---|
| Tested sequentially | Tested in parallel | All dual‐tested | ||||
| Number of patients | 791 | 372 | 150 | 169 | 319 | 844 |
| Mean age at referral (years) | 66.8 | 66.5 | 65.8 | 64.6 | 66.9 | 67.0 |
| SD | 10.2 | 9.6 | 9.4 | 8.3 | 10.2 | 10.2 |
| Number of patients with a pathogenic variant detected | 73 | 44 | 17 | 25 | 42 | 98 |
| % | 9.2% | 11.8% | 11.3% | 14.8% | 13.2% | 11.6% |
| 95% confidence interval | (7.2%, 11.2%) | (8.5%, 15.1%) | (6.3%, 16.4%) | (9.4%, 20.2%) | (9.5%, 16.9%) | (9.4%, 13.8%) |
An overlapping subset of both of these cohorts form part of the dual‐tested cohort.
Prevalence of pathogenic variants
Irrespective of the testing strategy, a pathogenic variant was detected in 11.6% (98/844) of patients. The pathogenic variants detected and the method of their detection are detailed in Table S1. Twelve variants were identified in more than one patient. The most common pathogenic variants were BRCA1 c.4065_4068del and BRCA1 c.2475del with each being identified in 4.1% (4/98) of those with a pathogenic variant. The prevalence of pathogenic variants according to testing strategy is detailed below and in Figure 2 and Table 3.
Germline testing
Germline testing was carried out in 791 patients. Of these, 319 patients also underwent a tumour test so were included in the dual‐tested cohort and the remaining 427 patients only underwent germline analysis. A germline pathogenic BRCA variant was detected in 73/791 (9.2%) patients; 60.3% (44/73) of variants were in BRCA1 and 39.7% (29/73) were in BRCA2. Two dosage variants (2.7% of all 73 germline variants) were identified: a BRCA1 deletion of exon 20 was found in one patient who only received a germline test and a duplication of BRCA1 exon 13 was found in one patient who underwent dual testing. The age of patients at the date of their referral to the AWMGS laboratory for testing was significantly younger in patients where a germline pathogenic BRCA variant was identified (mean age at referral 58.1 years, SD 9.4) compared with those where no germline pathogenic variant was detected (mean age at referral 67.7 years, SD 9.9) (Figure S1). Founder variants known to be of high prevalence in Ashkenazi Jewish populations were found in 2.7% (2/73) of patients with a germline pathogenic variant.
Tumour testing
Tumour testing was performed for 372 patients. At the time of this audit, 53 (14.2%) of the 372 patients that had received a tumour test had not yet had a germline test requested, but the majority (319/372, 85.8%) also had germline analysis so are included in the dual‐testing cohort. A pathogenic variant was detected in 11.8% (44/372) tumour samples. Two relatively large deletions, BRCA1 c.1607_1632del and BRCA2 c.4393_4438del, of 26 and 46 bases respectively, were identified through tumour testing.
Dual testing
Of the 319 patients that had both a germline and tumour BRCA test, 42 (13.2%) were found to have a pathogenic variant in the tumour and/or germline. Twenty (47.6%) of these pathogenic variants were germline and the additional 22 (52.4%) pathogenic variants were only identified through tumour testing. Therefore, 6.9% (22/319) of dual‐tested individuals had an acquired somatic variant. Eighteen (81.8%) of the 22 somatic variants were in BRCA1 and four (18.2%) were in BRCA2. One dosage variant (2.4% of 42 pathogenic variants), a duplication of BRCA1 exon 13, was only detected through germline analysis and not by tumour testing. The methods of detection of the pathogenic variants identified in the dual‐tested cohort are shown in Figure 3A.
Figure 3.
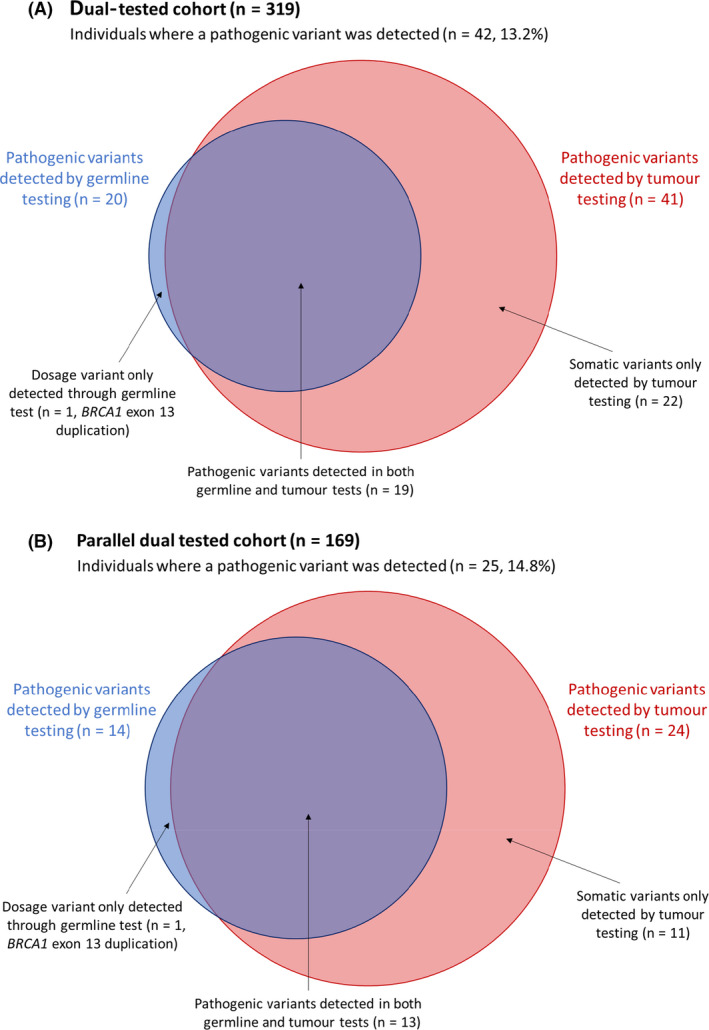
Method of detection of pathogenic BRCA1/2 variants identified in dual‐tested patients (A) and in parallel dual‐tested patients (B).
A total of 150 (47.0%) of the 319 patients that had dual testing received germline and somatic screening sequentially, where the result of the first test was issued before the second test being requested. The germline result was reported before the arrival of the tumour sample in the majority of these (86.7%, 130/150) in keeping with historical referral criteria. The mean time between the reporting of the germline result and the initiation of the tumour sample was 371.3 days (SD 347.6).
Parallel dual testing
A total of 169 individuals received a germline and tumour BRCA test in parallel and a pathogenic variant was detected in 25 (14.8%) of them. Fourteen of the 25 pathogenic variants (56.0%) identified were present in the germline. The additional 11/25 (44.0%) variants were only detected in the tumour, so acquired somatic variants were found in 11/169 (6.5%) of the parallel dual‐tested cohort, with eight (72.7%) of these in BRCA1 and three (27.3%) in BRCA2. As was the case in the full dual‐tested cohort, tumour testing was able to detect the majority (24/25, 96.0%) of the pathogenic variants with one dosage variant (1/25, 4.0%) not detected on a tumour test. The methods of detection of the pathogenic variants identified in the parallel dual‐tested cohort are shown in Figure 3(B). The mean time between the arrival of the first sample and when the results for both tests were available was 48.9 days (SD 19.5).
Discussion
Main findings
This is one of the largest UK studies to publish data examining the outcomes of BRCA genetic testing in patients with high‐grade serous ovarian cancer and the first to report outcomes from parallel germline and tumour testing. The AWMGS has been offering germline and tumour BRCA testing for this patient population in Wales since 2015 and 2017, respectively, through an oncology‐led testing pathway. This mainstreamed approach allows the large‐scale delivery of testing to patients who may rely on a rapid genetic result to guide treatment decisions. It has been shown to be effective in other centres and is now recommended in the British Gynaecological Cancer Society national guidelines. 19
Germline pathogenic BRCA variant detection
The prevalence of germline BRCA pathogenic variants in this study was 9.2% (73/791), with more variants reported in BRCA1 than BRCA2 (44, 60.3% versus 29, 39.7%, respectively), and only 2.7% of patients (2/73 of those with germline variants) had BRCA founder variants associated with an Ashkenazi Jewish ancestry. The observed prevalence of germline pathogenic variants was consistent with a study conducted in a largely homogeneous population with regards to ethnicity that also lacked founder variants. 13 However it was lower than several reports in the literature from other centres (13–18.5%) with higher rates of founder variants (11–17.6%) that included more diverse tumour types (Table 1). 10 , 11 Wales was the least ethnically diverse area according to the Office for National Statistics 2011 Census data for England and Wales, and we expect the ethnicities of the individuals included in this study to be in‐line with this. We therefore consider it reasonable to suggest that tumour type, the prevalence of founder variants and variation in the ethnic composition of patients may have an association with germline BRCA variant detection. Furthermore, germline pathogenic BRCA variants, are known to be associated with individuals with a strong family history of BRCA‐related cancers. According to reports in the literature, around half (45.5–53.2%) of ovarian cancer patients with BRCA pathogenic variants have been shown to have a significant family history. 12 , 23 In Wales, these patients with ovarian cancer and a family history or secondary personal diagnosis of breast and/or ovarian cancer may have been referred for germline BRCA testing through the clinical genetics service, not through the oncologist‐led pathway. 18 It is therefore possible to assume that this study may not have captured the data from some of these individuals with a strong family history of BRCA‐related cancers, who are more likely to have a germline BRCA pathogenic variant. It will be interesting to see the outcomes of larger UK studies, such as the SIGNPOST study, which may be able to fully elucidate the correlation between family history, founder variant rate, ethnicity and/or tumour type with germline pathogenic BRCA variant detection rate, as has been suggested by this audit and other regional studies.
Paired germline and tumour testing
The pathogenic variant pick‐up rate was 13.2% (42/319) for patients who underwent both a germline and a tumour BRCA genetic test. However, before 2019, BRCA tumour testing was only available to patients with a germline normal result. The referral of patients for a germline and tumour test through this strategy is therefore not independent and the whole dual‐tested cohort may be enriched for germline normal patients (as illustrated in Figure 4). To remove bias from this dual‐tested cohort, the outcomes from patients who received germline and tumour analysis in parallel (where testing overlapped and referral for each test was therefore independent) were examined. There were 169 patients in this concurrently tested cohort and the pathogenic BRCA variant rate was 14.8% (25/169). This is higher than either of the single‐test strategies as pathogenic dosage and acquired variants would both be detectable through dual testing but would be missed using a tumour‐only or germline‐only testing strategy, respectively. It is also higher than in the cohort where dual testing was not independent, as expected, because individuals with a germline pathogenic variant, that may have not progressed to have a tumour test based on historical referral criteria, would still be included in the parallel‐tested group (Figure 4). An acquired somatic BRCA variant (not present in the germline) was detected in 11/169 (6.5%) of concurrently dual‐tested patients in this study with the majority (72.7%, 8/11) of the variants identified in BRCA1. This is in‐line with other studies (Table 2) that have reported similar somatic variant detection rates of 4.1–7.1%. 15 , 16 , 17 The acquired somatic variant rate represents a significant proportion of patients that are eligible for PARPi treatment that would not be identified through germline screening alone and highlights the importance of access to tumour testing services for this population. Furthermore, access to PARPi treatment may have been delayed for these individuals if the historical two‐stage referral pathway was employed, highlighting the value of carrying out parallel testing to ensure timely access to the most appropriate treatment for patients with high‐grade serous ovarian cancer.
Figure 4.
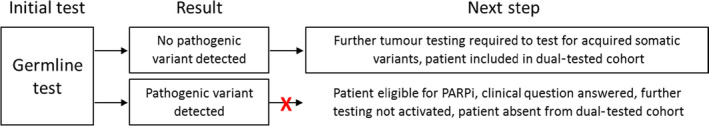
Potential source of bias in dual‐tested cohort due to historical two‐step referral criteria.
Dosage variants
Two BRCA dosage variants (constituting 2.7% of all 73 germline variants) were detected during germline testing in this study: a recurrent and well‐documented BRCA1 exon 13 duplication 24 and a BRCA1 exon 20 deletion. This means a possible 2.0% (2/98) of patients found to have a pathogenic variant in this study would not have been eligible for PARPi if a tumour testing‐only strategy was followed. Dosage variants have been reported to represent 5.9–12.1% of germline pathogenic variants in other studies (Table 1), but also in a low number of patients, and unpublished data from the SIGNPOST study and evidence from Public Health England suggest that this could be as much as 10% or more in various populations that have undergone BRCA genetic screening. 4 , 11 , 12 , 19 Given the astonishing potential clinical benefit for patients who receive PARPi treatment and the implications for family members of those with germline pathogenic variants, it would therefore not be appropriate to follow a tumour‐testing‐only strategy and risk not identifying patients/families harbouring large rearrangements in the BRCA genes. 25
Limitations
Information on screening uptake, patient satisfaction, tumour characteristics, family history, ethnicity, previous treatment regimens, response and follow up was not within the scope of this retrospective audit and has not been examined.
The region of interest examined in the BRCA genes in this study is limited to the coding exons and immediate flanking intronic regions that are covered to a minimum read depth threshold. Therefore, variants that lie outside this region of interest or that fall in regions not covered by sufficient read depths would not be detected by either tumour or germline BRCA testing.
Future directions
The indication for the use of PARPi is rapidly evolving. All patients with high‐grade, non‐mucinous ovarian cancer in Wales are now eligible for BRCA testing, as is done in other NHS laboratories, and some types of PARPi will soon be accessible to all these patients regardless of BRCA status. The testing strategy in Wales is also due to expand to include concurrent BRCA and genomic instability testing using a tumour sample alongside germline BRCA testing to help assess how well these patients may benefit from PARPi treatment and which PARPi to consider. The extension of testing strategies to include other homologous recombination repair genes may also become relevant considering the active research ongoing into the use of PARPi for cancer patients with pathogenic variants in genes such as PALB2. 26 , 27
The results of this study will be important for informing on the optimal testing strategies for other disease areas that may employ paired germline and tumour testing in the future to identify patients eligible for various treatments. For instance, the utility of PARPi in BRCA‐positive patients with prostate cancer or breast cancer is currently being investigated in the UK. Furthermore, the application of NGS and bioinformatics technologies that have the capability to detect large genomic rearrangements in tumour samples is on the horizon in routine testing in NHS laboratories and will make significant strides towards the streamlining of this testing pathway.
Conclusions
In this study, parallel tumour and germline testing was carried out for 169 patients with high‐grade serous ovarian cancer and pathogenic variants were detected at a frequency of 14.8%, with 6.5% of patients shown to have an acquired pathogenic variant that would not be detected through germline testing alone. Germline NGS techniques can identify large dosage variants that are often undetectable by the NGS methods currently employed to test tumour tissue. Consequently, this study identified a possible 2.0% of patients with a pathogenic variant that would not have been eligible for PARPi if a tumour‐only testing strategy was followed.
Based on the findings from this national study, we suggest that the availability of germline and tumour BRCA1 and BRCA2 NGS testing services, requested in parallel, would ensure that all eligible patients with non‐mucinous ovarian, fallopian tube or peritoneal cancer are afforded the opportunity to access the significant benefits of PARPi treatment in a timely way.
Disclosure of interests
Full disclosure of interests forms are available to view online as supporting information.
Contribution to authorship
The study was designed by SM, RW and SPS. Data were acquired by BF, AM, RJ, LH, CF, EH, AM and AQ. Data analysis and manuscript preparation were by BF. All authors reviewed the manuscript before submission.
Details of ethics approval
Ethical approval was not needed to undertake this audit. Where appropriate, all principles of the Declaration of Helsinki were adhered to.
Funding
No external funding was received specifically for this work.
Acknowledgements
The authors would like to thank all staff in the genetics, oncology and pathology services in Wales that contributed to this work.
Data availability
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available because of privacy or ethical restrictions.
Consent
Consent for testing was obtained by the referring oncologist. No patient identifying information has been disclosed in this audit.
Supporting information
Figure S1. Age of patients at date of referral to AWMGS by germline BRCA1/2 testing outcome. Tukey box plot; shown are minimum, lower quartile, median, upper quartile and maximum ages for each data set. Mean age at referral 58.1 years (SD 9.4) for group with a germline pathogenic variant identified versus 67.7 years (SD 9.9) for the group with no pathogenic germline variant identified.
Table S1. Pathogenic BRCA1 and BRCA2 variants identified in this study according to testing strategy.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Frugtniet B, Morgan S, Murray A, Palmer‐Smith S, White R, Jones R, Hanna L, Fuller C, Hudson E, Mullard A, Quinton AE. The detection of germline and somatic BRCA1/2 genetic variants through parallel testing of patients with high‐grade serous ovarian cancer: a national retrospective audit. BJOG 2022; 129:433–442.
References
- 1. Ray‐Coquard I, Pautier P, Pignata S, Pérol D, González‐Martín A, Berger R, et al. Olaparib plus bevacizumab as first‐line maintenance in ovarian cancer. N Engl J Med 2019;381:2416–28. [DOI] [PubMed] [Google Scholar]
- 2. Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in patients with platinum‐sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol 2014;15:852–61. [DOI] [PubMed] [Google Scholar]
- 3. Oza AM, Tinker AV, Oaknin A, Shapira‐Frommer R, McNeish IA, Swisher EM, et al. Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high‐grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: integrated analysis of data from Study 10 and ARIEL2. Gynecol Oncol 2017;147:267–75. [DOI] [PubMed] [Google Scholar]
- 4. Moore K, Colombo N, Scambia G, Kim B‐G, Oaknin A, Friedlander M, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2018;379:2495–505. [DOI] [PubMed] [Google Scholar]
- 5. Coleman RL, Oza AM, Lorusso D, Aghajanian C, Oaknin A, Dean A, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet 2017;390:1949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mohyuddin GR, Aziz M, Britt A, Wade L, Sun W, Baranda J, et al. Similar response rates and survival with PARP inhibitors for patients with solid tumors harboring somatic versus Germline BRCA mutations: a meta‐analysis and systematic review. BMC Cancer 2020;20:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuchenbaecker KB, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 2017;317:2402–16. [DOI] [PubMed] [Google Scholar]
- 8. Lewis KE, Lu K, Klimczak A, Mok S. Recommendations and choices for BRCA mutation carriers at risk for ovarian cancer: a complicated decision. Cancers 2018;10:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rust K, Spiliopoulou P, Tang CY, Bell C, Stirling D, Phang T, et al. Routine germline BRCA1 and BRCA2 testing in patients with ovarian carcinoma: analysis of the Scottish real‐life experience. BJOG 2018;125:1451–8. [DOI] [PubMed] [Google Scholar]
- 10. Rahman B, Lanceley A, Kristeleit RS, Ledermann JA, Lockley M, McCormack M, et al. Mainstreamed genetic testing for women with ovarian cancer: first‐year experience. J Med Genet 2019;56:195–8. [DOI] [PubMed] [Google Scholar]
- 11. Rumford M, Lythgoe M, McNeish I, Gabra H, Tookman L, Rahman N, et al. Oncologist‐led BRCA 'mainstreaming' in the ovarian cancer clinic: a study of 255 patients and its impact on their management. Sci Rep 2020;10:3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. George A, Riddell D, Seal S, Talukdar S, Mahamdallie S, Ruark E, et al. Implementing rapid, robust, cost‐effective, patient‐centred, routine genetic testing in ovarian cancer patients. Sci Rep 2016;6:29506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Plaskocinska I, Shipman H, Drummond J, Thompson E, Buchanan V, Newcombe B, et al. New paradigms for BRCA1/BRCA2 testing in women with ovarian cancer: results of the Genetic Testing in Epithelial Ovarian Cancer (GTEOC) study. J Med Genet 2016;53:655–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flaum N, Morgan RD, Burghel GJ, Bulman M, Clamp AR, Hasan J, et al. Mainstreaming germline BRCA1/2 testing in non‐mucinous epithelial ovarian cancer in the North West of England. Eur J Hum Genet 2020;28:1541–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koczkowska M, Zuk M, Gorczynski A, Ratajska M, Lewandowska M, Biernat W, et al. Detection of somatic BRCA1/2 mutations in ovarian cancer ‐ next‐generation sequencing analysis of 100 cases. Cancer Med 2016;5:1640–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eoh KJ, Kim HM, Lee J‐Y, Kim S, Kim SW, Kim YT, et al. Mutation landscape of germline and somatic BRCA1/2 in patients with high‐grade serous ovarian cancer. BMC Cancer 2020;20:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peixoto A, Pinto P, Guerra J, Pinheiro M, Santos C, Pinto C, et al. Tumor testing for somatic and germline BRCA1/BRCA2 variants in ovarian cancer patients in the context of strong founder effects. Front Oncol 2020;10:1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kansu B, Gardner J, Price‐Tate R, Murch O, Murray A. BRCA gene testing in women with high‐grade serous ovarian carcinoma. J Obstet Gynaecol 2020;41:962–5. [DOI] [PubMed] [Google Scholar]
- 19. Sundar S, Manchanda R, Gourley C, George A, Wallace A, Balega J, et al. British Gynaecological Cancer Society/British Association of Gynaecological Pathology consensus for germline and tumor testing for BRCA1/2 variants in ovarian cancer in the United Kingdom. Int J Gynecol Cancer 2021;31:272–8. [DOI] [PubMed] [Google Scholar]
- 20. Ellard S, Baple EL, Berry I. ACGS best practice guidelines for variant classification in rare disease 2020. Assoc Clin Genomic Sci (ACGS) 2020:1–33. https://www.acgs.uk.com/quality/best‐practice‐guidelines/ [Google Scholar]
- 21. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier‐Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garrett A, Callaway A, Durkie M, Cubuk C, Alikian M, Burghel GJ, et al. Cancer Variant Interpretation Group UK (CanVIG‐UK): an exemplar national subspecialty multidisciplinary network. J Med Genet 2020;57:829–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alsop K, Fereday S, Meldrum C, deFazio A, Emmanuel C, George J, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation‐positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol 2012;30:2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. The BRCA Exon Duplication Screening Group . The exon 13 duplication in the BRCA1 gene is a founder mutation present in geographically diverse populations. The BRCA1 Exon 13 Duplication Screening Group. Am J Hum Genet 2000;67:207–12. [PMC free article] [PubMed] [Google Scholar]
- 25. Konstantinopoulos PA, Norquist B, Lacchetti C, Armstrong D, Grisham RN, Goodfellow PJ, et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J Clin Oncol 2020;38:1222–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grellety T, Peyraud F, Sevenet N, Tredan O, Dohollou N, Barouk‐Simonet E, et al. Dramatic response to PARP inhibition in a PALB2‐mutated breast cancer: moving beyond BRCA. Ann Oncol 2020;31:822–3. [DOI] [PubMed] [Google Scholar]
- 27. Kuemmel S, Harrach H, Schmutzler RK, Kostara A, Ziegler‐Löhr K, Dyson MH, et al. Olaparib for metastatic breast cancer in a patient with a germline PALB2 variant. NPJ Breast Cancer 2020;6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Age of patients at date of referral to AWMGS by germline BRCA1/2 testing outcome. Tukey box plot; shown are minimum, lower quartile, median, upper quartile and maximum ages for each data set. Mean age at referral 58.1 years (SD 9.4) for group with a germline pathogenic variant identified versus 67.7 years (SD 9.9) for the group with no pathogenic germline variant identified.
Table S1. Pathogenic BRCA1 and BRCA2 variants identified in this study according to testing strategy.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available because of privacy or ethical restrictions.


