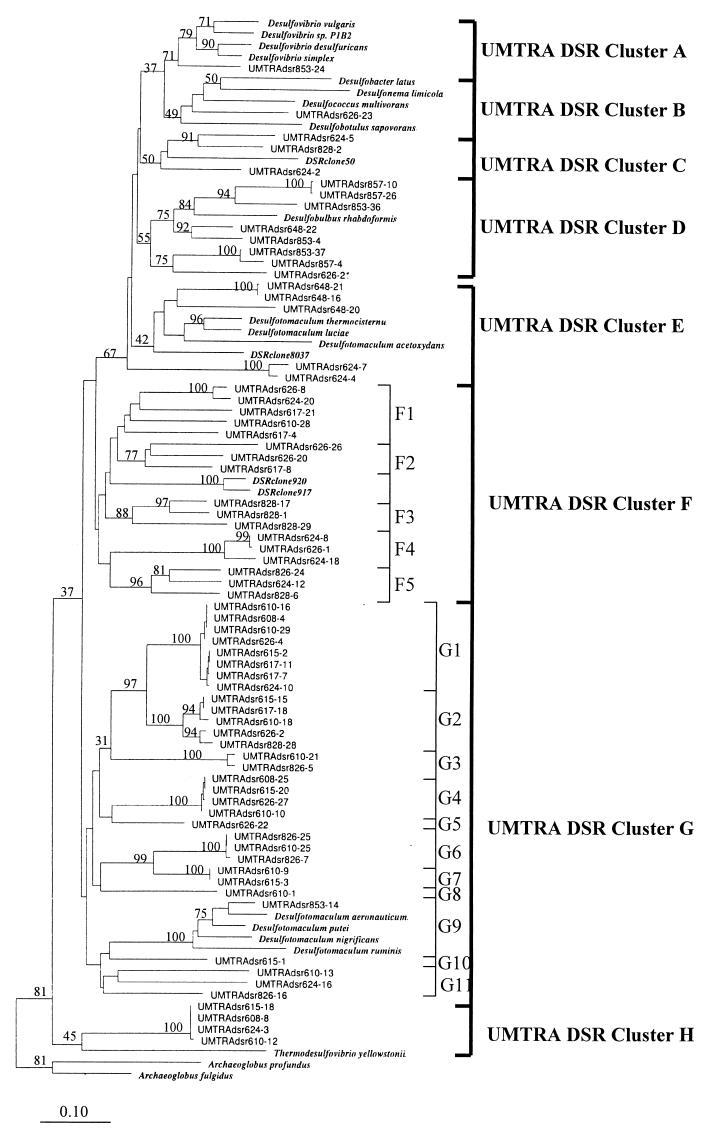
FIG. 4.
Neighbor-joining analysis of DSR α-subunit fragments selected from clone libraries as frequently occurring MspI digestion patterns. Sequences prefixed “UMTRAdsr” were generated during this study. The prefix is followed by the sample well number, which is followed by the clone number. Clones were selected from libraries on the basis of MspI patterns to provide a preliminary survey of the most commonly recovered DSR gene sequences from each sample. Nucleotide sequence accession numbers are given in parentheses for the following organisms: Desulfobulbus rhabdoformis (AJ250473), Desulfovibrio desulfuricans (AJ289157), Desulfococcus multivorans (AJ277107), Desulfotomaculum thermocisternum (F074396), Archaeoglobus profundus (AF071499), Thermodesulfovibrio yellowstonii (U58122), Desulfobotulus sapovorans (U58120), Desulfovibrio sp. strain P1B2 (U58116), Desulfotomaculum ruminis (U58118), Desulfococcus multivorans (U58126), Desulfonema limicola (U58128), Desulfobacter latus (U58124), Desulfovibrio vulgaris (U16723), Archaeoglobus fulgidus (M95624), and Desulfovibrio simplex (U78738). Gene sequences from Desulfotomaculum luciae, D. acetoxidans, D. putei, D. aeronauticum, D. nigrificans, and Desulfovibrio desulfuricans were generated in this study under accession no. AY015493 to AY015499. The numbers on the tree refer to bootstrap values on 100 replicates; only values above 30 are given. Scale bar represents 10% estimated change.