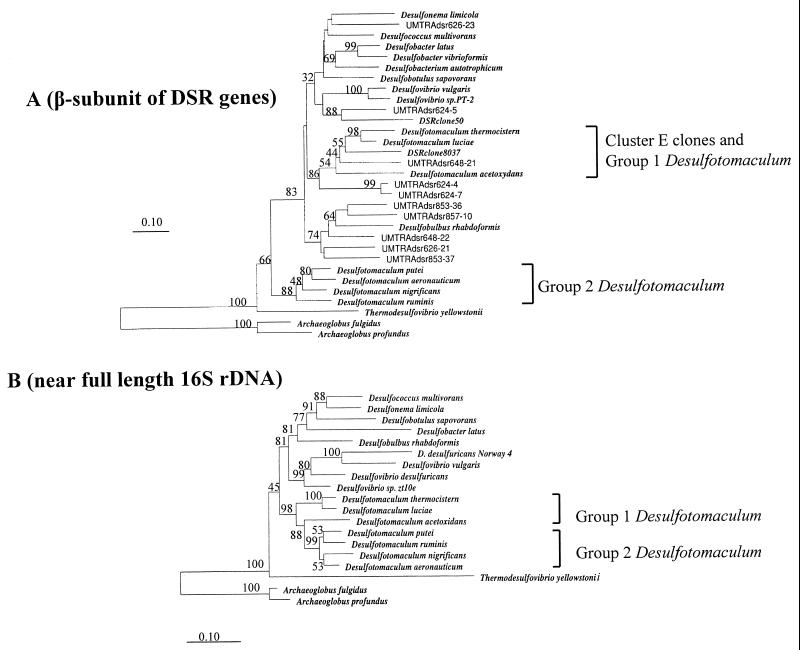
FIG. 5.
(A) Neighbor-joining analysis of some DSR β-subunit sequences from UMTRA Shiprock site clones and reference pure culture organisms. Sequences prefixed “UMTRAdsr” were generated during this study. Nucleotide sequence accession numbers are given in parentheses for the following organisms: Desulfobulbus rhabdoformis (AJ250473), Desulfococcus multivorans (U58127), Desulfotomaculum thermocisternum (AF074396), Archaeoglobus profundus (AF071499), Thermodesulfovibrio yellowstonii (U58123), Desulfobotulus sapovorans (U58121), Desulfovibrio sp. strain PT-2. (U58115), Desulfotomaculum ruminis (U58119), Desulfonema limicola (U58129), Desulfobacter latus (U58125), Desulfovibrio vulgaris (U16723), Archaeoglobus fulgidus (M95624), Desulfobacter vibrioformis (AJ250472), Desulfobacterium autotrophicum (Y15478), DSR clone 50 (AF179329), and DSR clone 8037 (AF179336). Gene sequences from Desulfotomaculum luciae, D. acetoxidans, D. putei, D. aeronauticum, and D. nigrificans were generated in this study under accession no. AY015577 to AY015581. (B) Neighbor-joining analysis of near full-length 16S rDNA from pure cultures. Nucleotide sequence accession numbers are given in parentheses for the following organisms: Desulfotomaculum ruminis (Y11572), D. putei strain SMCCW 459 (AF053929), D. aeronauticum (X98407), D. acetoxidans (Y11566), D. thermocisternum (U33455), D. nigrificans (X62176), D. luciae (AF069293), Desulfococcus multivorans ATCC 33890 (M34405), Desulfonema limicola (U45990), Desulfobacter latus (M34414), Desulfobotulus sapovorans (M34402), Desulfovibrio vulgaris (M34399), D. desulfuricans (AF098671), Archaeoglobus profundus. (AF272841), Desulfovibrio sp. strain zt10e (AF109469), Archaeoglobus fulgidus (Y00275), and D. desulfuricans sp. strain Norway 4 (M37312). The numbers on the trees refer to bootstrap values on 100 replicates; only values above 30 are given. The scale bar represents 10% estimated change.