Fig. 4.
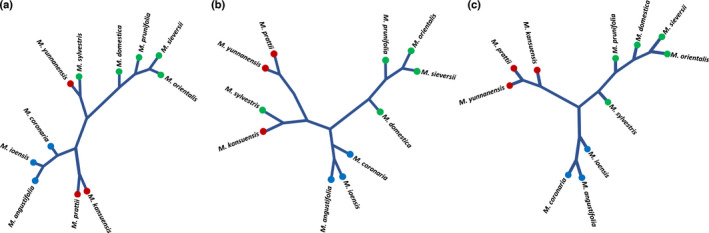
Results of hierarchical clustering based on Bray–Curtis dissimilarity distances of the fungal (a) and bacterial (b) community composition using clustering method ‘average’. (c) Shows the phylogenetic tree based on Malus ITS gene. Malus phylogenetic distance was inferred by using the neighbor‐joining tree estimation in R package phangorn. The leaf color indicates Malus groups: green = Malus × domestica and its wild progenitors (M. sieversii, M. orientalis, M. prunifolia, and M. sylvestris), blue = North American species (M. angustifolia, M. coronaria, and M. ioensis), and red = Asian species (M. kansuensis, M. yunnanensis, and M. prattii). The phylogenic plots were visualized using the fviz_dend function in the R package factoextra v.1.0.7.
