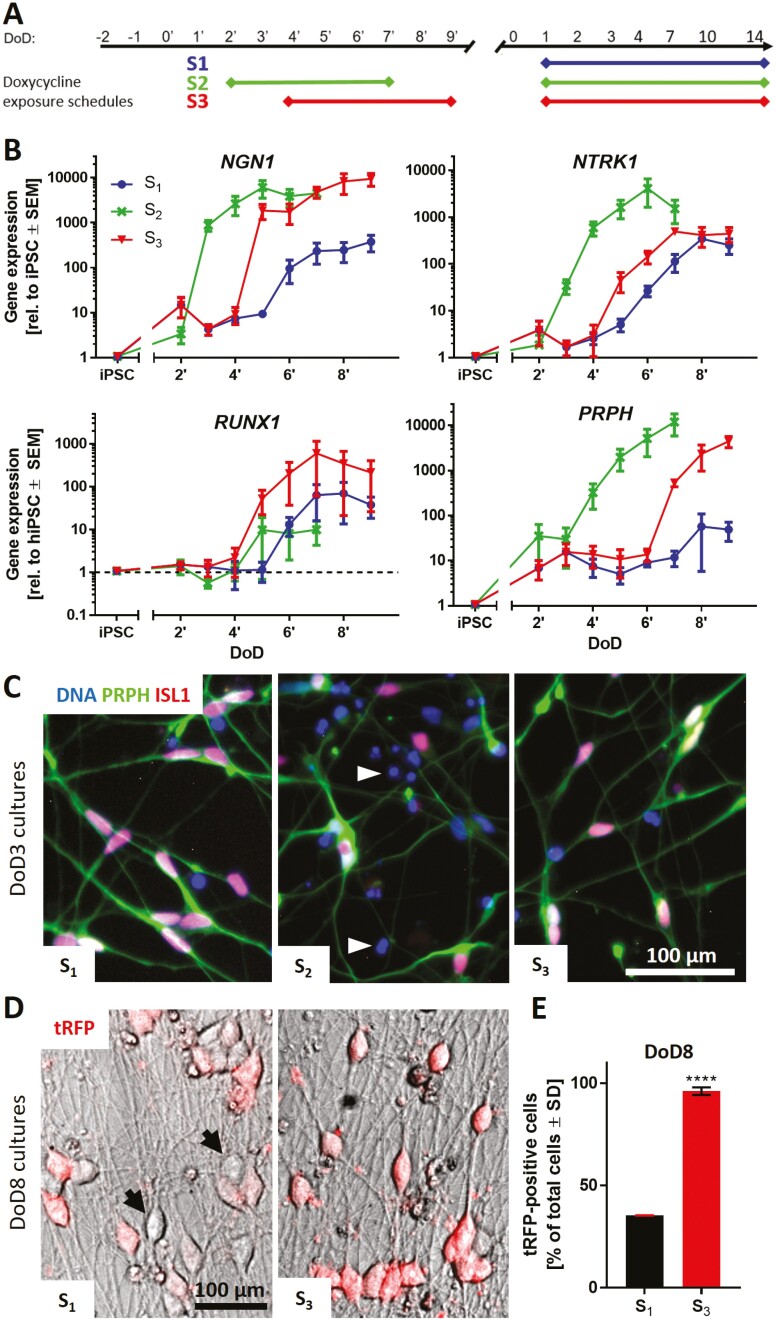
Figure 3.
Integration of NGN1-overexpression in the standard small molecule differentiation protocol. (A) Schematic representation of 3 different doxycycline exposure schedules (S1, S2, S3) incorporated in the standard differentiation protocol. Condition S1 did not receive doxycycline treatment before freezing. Cells of condition S2 were treated with doxycycline from DoD2ʹ until DoD7ʹ with subsequent freezing of the cells. Condition S3 included doxycycline treatment from DoD4ʹ until DoD9ʹ with subsequent freezing. After thawing, all 3 conditions were exposed to doxycycline from DoD1 until DoD14. (B) Gene expression analysis of the nociceptor marker genes NGN1, NTRK1, RUNX1 and the general sensory neuronal marker gene peripherin (PRPH) for all 3 exposure situations. Data are expressed relative to expression levels in iPSCs and given as means ± SEM of 3-4 biological replicates. (C) Immunofluorescence images of cultures (S1-3) on DoD3 (after thawing). Cells were stained for peripherin (PRPH) and ISL1. Nuclei were stained with H33342 (DNA). Details are displayed in Supplementary Fig. S4A. White arrowheads indicate exemplary dead cells. (D) Overlay of phase contrast and tRFP fluorescence images of condition S1 and S3 neuronal cultures on DoD8. Black arrows indicate exemplary tRFP-negative cells. (E) Quantification of tRFP positive cells in cultures of differentiation conditions S1 and S3 on DoD8. Data are shown as percentage of total cell count ± SD. DoD, day of differentiation; tRFP, turbo red fluorescent protein.