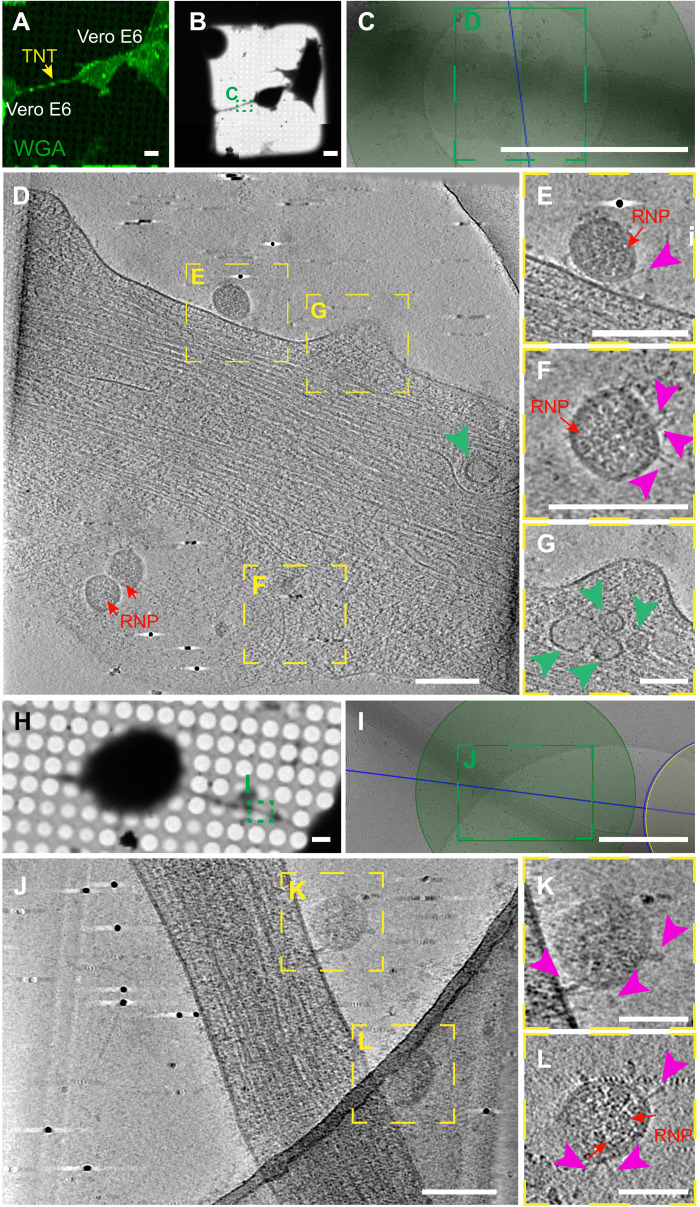
Fig. 6. Cryo-CLEM reveal SARS-CoV-2 on TNTs between infected Vero E6 cells.
(A) TNT-connected SARS-CoV-2–infected Vero E6 cells stained with wheat germ agglutinin (WGA) (green) and acquired by confocal microscopy (A), with low (B) and intermediate (C) magnification TEM. (D) Slices of tomograms of TNT in green dashed square in (C) showing the extracellular SARS-CoV-2 on top of the TNT connecting Vero E6 cells. (E to G) High-magnification cryo-ET slices corresponding to the yellow dashed squares in (D) showing SARS-CoV-2; RNPs and S proteins are observed. Pink arrowheads indicate the spike; red arrows point the RNPs. (E and F) High-magnification cryo-ET slices showing the extracellular virions on TNT. (G) High-magnification cryo-ET slices corresponding to the yellow dashed squares in (D). (G) The slices tomograms showed small vesicle compartments with a diameter of around 50 to 100 nm inside TNT. Green arrowheads point the vesicles. (H and I) Low (H) and intermediate (I) magnification of an electron micrograph of TNT-connected SARS-CoV-2–infected Vero E6 cells. (J) High-magnification cryo-ET slice corresponding to the green dashed rectangle in (I). (K and L) High-magnification cryo-ET slices showing the extracellular virions on TNT. Scale bars, 10 μm (A, B, and H), 2 μm (C), 150 nm (D to F and J), 100 nm (G, K, and L), and 1 μm (I).